当前位置:
X-MOL 学术
›
Curr. Genomics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
iHyd-LysSite (EPSV): Identifying Hydroxylysine Sites in Protein Using Statistical Formulation by Extracting Enhanced Position and Sequence variant Feature Technique
Current Genomics ( IF 1.8 ) Pub Date : 2020-10-22 , DOI: 10.2174/1389202921999200831142629 Muhammad Khalid Mahmood 1 , Asma Ehsan 1 , Yaser Daanial Khan 1 , Kuo-Chen Chou 1
Current Genomics ( IF 1.8 ) Pub Date : 2020-10-22 , DOI: 10.2174/1389202921999200831142629 Muhammad Khalid Mahmood 1 , Asma Ehsan 1 , Yaser Daanial Khan 1 , Kuo-Chen Chou 1
Affiliation
|
Introduction
Hydroxylation is one of the most important post-translational modifications (PTM) in cellular functions and is linked to various diseases. The addition of one of the hydroxyl groups (OH) to the lysine sites produces hydroxylysine when undergoes chemical modification. Methods
The method which is used in this study for identifying hydroxylysine sites based on powerful mathematical and statistical methodology incorporating the sequence-order effect and composition of each object within protein sequences. This predictor is called "iHyd-LysSite (EPSV)" (identifying hydroxylysine sites by extracting enhanced position and sequence variant technique). The prediction of hydroxylysine sites by experimental methods is difficult, laborious and highly expensive. In silico technique is an alternative approach to identify hydroxylysine sites in proteins. Results
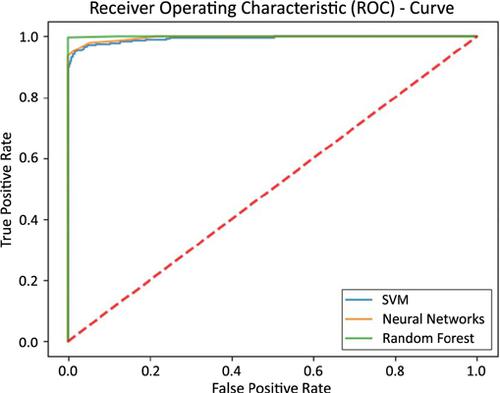
The experimental results require that the predictive model should have high sensitivity and specificity values and must be more accurate. The self-consistency, independent, 10-fold cross-validation and jackknife tests are performed for validation purposes. These tests are resulted by using three renowned classifiers, Neural Networks (NN), Random Forest (RF) and Support Vector Machine (SVM) with the demanding prediction rate. The overall predictive outcomes are extraordinarily superior to the results obtained by previous predictors. The proposed model contributed an excellent prediction rate in the system for NN, RF, and SVM classifiers. The sensitivity and specificity results using all these classifiers for jackknife test are 96.08%, 94.99%, 98.16% and 97.52%, 98.52%, 80.95%. Conclusion
The results obtained by the proposed tool show that this method may meet the future demand of hydroxylysine sites with a better prediction rate over the existing methods.
中文翻译:

iHyd-LysSite (EPSV):通过提取增强的位置和序列变体特征技术,使用统计公式识别蛋白质中的羟基赖氨酸位点
引言 羟基化是细胞功能中最重要的翻译后修饰 (PTM) 之一,与各种疾病有关。当进行化学修饰时,向赖氨酸位点添加一个羟基 (OH) 会产生羟基赖氨酸。方法 本研究中使用的方法是基于强大的数学和统计方法来识别羟赖氨酸位点,该方法结合了序列顺序效应和蛋白质序列中每个对象的组成。该预测器称为“iHyd-LysSite (EPSV)”(通过提取增强的位置和序列变异技术来识别羟赖氨酸位点)。通过实验方法预测羟赖氨酸位点是困难的、费力的和非常昂贵的。计算机技术是识别蛋白质中羟赖氨酸位点的另一种方法。结果 实验结果要求预测模型应具有较高的敏感性和特异性值,并且必须更加准确。出于验证目的,执行自洽、独立、10 折交叉验证和折刀测试。这些测试是通过使用三个著名的分类器产生的,神经网络 (NN)、随机森林 (RF) 和支持向量机 (SVM) 具有苛刻的预测率。总体预测结果非常优于先前预测器获得的结果。所提出的模型在系统中为 NN、RF 和 SVM 分类器提供了出色的预测率。使用所有这些分类器进行折刀试验的敏感性和特异性结果分别为 96.08%、94.99%、98。16% 和 97.52%、98.52%、80.95%。结论 所提出的工具获得的结果表明,该方法可以满足未来对羟赖氨酸位点的需求,并且比现有方法具有更好的预测率。
更新日期:2020-10-22
中文翻译:

iHyd-LysSite (EPSV):通过提取增强的位置和序列变体特征技术,使用统计公式识别蛋白质中的羟基赖氨酸位点
引言 羟基化是细胞功能中最重要的翻译后修饰 (PTM) 之一,与各种疾病有关。当进行化学修饰时,向赖氨酸位点添加一个羟基 (OH) 会产生羟基赖氨酸。方法 本研究中使用的方法是基于强大的数学和统计方法来识别羟赖氨酸位点,该方法结合了序列顺序效应和蛋白质序列中每个对象的组成。该预测器称为“iHyd-LysSite (EPSV)”(通过提取增强的位置和序列变异技术来识别羟赖氨酸位点)。通过实验方法预测羟赖氨酸位点是困难的、费力的和非常昂贵的。计算机技术是识别蛋白质中羟赖氨酸位点的另一种方法。结果 实验结果要求预测模型应具有较高的敏感性和特异性值,并且必须更加准确。出于验证目的,执行自洽、独立、10 折交叉验证和折刀测试。这些测试是通过使用三个著名的分类器产生的,神经网络 (NN)、随机森林 (RF) 和支持向量机 (SVM) 具有苛刻的预测率。总体预测结果非常优于先前预测器获得的结果。所提出的模型在系统中为 NN、RF 和 SVM 分类器提供了出色的预测率。使用所有这些分类器进行折刀试验的敏感性和特异性结果分别为 96.08%、94.99%、98。16% 和 97.52%、98.52%、80.95%。结论 所提出的工具获得的结果表明,该方法可以满足未来对羟赖氨酸位点的需求,并且比现有方法具有更好的预测率。











































 京公网安备 11010802027423号
京公网安备 11010802027423号