Current Analytical Chemistry ( IF 1.7 ) Pub Date : 2020-11-30 , DOI: 10.2174/1573411016999200518083359 Nafiseh Vahedi 1 , Majid Mohammad Hosseini 1 , Mehdi Nekoei 1
|
Background: The poly(ADP-ribose) polymerases (PARP) is a nuclear enzyme superfamily present in eukaryotes.
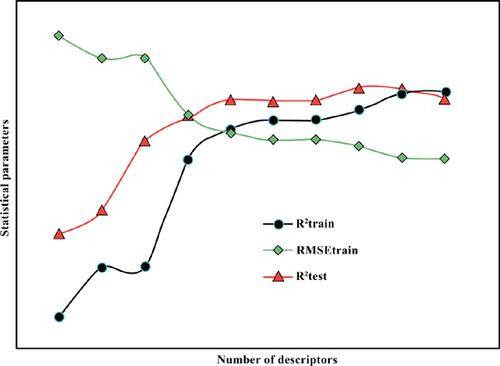
Methods: In the present report, some efficient linear and non-linear methods including multiple linear regression (MLR), support vector machine (SVM) and artificial neural networks (ANN) were successfully used to develop and establish quantitative structure-activity relationship (QSAR) models capable of predicting pEC50 values of tetrahydropyridopyridazinone derivatives as effective PARP inhibitors. Principal component analysis (PCA) was used to a rational division of the whole data set and selection of the training and test sets. A genetic algorithm (GA) variable selection method was employed to select the optimal subset of descriptors that have the most significant contributions to the overall inhibitory activity from the large pool of calculated descriptors.
Results: The accuracy and predictability of the proposed models were further confirmed using crossvalidation, validation through an external test set and Y-randomization (chance correlations) approaches. Moreover, an exhaustive statistical comparison was performed on the outputs of the proposed models. The results revealed that non-linear modeling approaches, including SVM and ANN could provide much more prediction capabilities.
Conclusion: Among the constructed models and in terms of root mean square error of predictions (RMSEP), cross-validation coefficients (Q2 LOO and Q2 LGO), as well as R2 and F-statistical value for the training set, the predictive power of the GA-SVM approach was better. However, compared with MLR and SVM, the statistical parameters for the test set were more proper using the GA-ANN model.
中文翻译:

GA-MLR,GA-SVM和GA-ANN方法对PARP抑制剂的QSAR研究
背景:聚(ADP-核糖)聚合酶(PARP)是真核生物中存在的核酶超家族。
方法:在本报告中,成功地使用了一些有效的线性和非线性方法,包括多元线性回归(MLR),支持向量机(SVM)和人工神经网络(ANN),以开发和建立定量构效关系(QSAR) )模型能够预测作为有效PARP抑制剂的四氢吡啶并吡啶并嗪酮衍生物的pEC50值。主成分分析(PCA)用于整个数据集的合理划分以及训练和测试集的选择。遗传算法(GA)变量选择方法用于从大量已计算的描述符中选择对总体抑制活性贡献最大的描述符的最佳子集。
结果:使用交叉验证,通过外部测试集验证和Y随机化(机会相关)方法,进一步证实了所提出模型的准确性和可预测性。此外,对提出的模型的输出进行了详尽的统计比较。结果表明,包括SVM和ANN在内的非线性建模方法可以提供更多的预测功能。
结论:在所构建的模型中,就预测的均方根误差(RMSEP),交叉验证系数(Q2 LOO和Q2 LGO)以及训练集的R2和F统计值而言,预测模型的预测能力GA-SVM方法更好。但是,与MLR和SVM相比,使用GA-ANN模型的测试集统计参数更为合适。











































 京公网安备 11010802027423号
京公网安备 11010802027423号