Cell Host & Microbe ( IF 20.6 ) Pub Date : 2020-10-28 , DOI: 10.1016/j.chom.2020.10.004 Daniel Bojar 1 , Rani K Powers 1 , Diogo M Camacho 2 , James J Collins 3
|
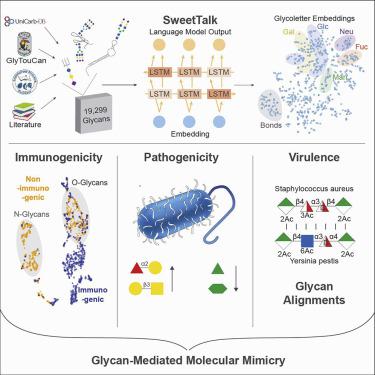
Glycans, the most diverse biopolymer, are shaped by evolutionary pressures stemming from host-microbe interactions. Here, we present machine learning and bioinformatics methods to leverage the evolutionary information present in glycans to gain insights into how pathogens and commensals interact with hosts. By using techniques from natural language processing, we develop deep-learning models for glycans that are trained on a curated dataset of 19,299 unique glycans and can be used to study and predict glycan functions. We show that these models can be utilized to predict glycan immunogenicity and the pathogenicity of bacterial strains, as well as investigate glycan-mediated immune evasion via molecular mimicry. We also develop glycan-alignment methods and use these to analyze virulence-determining glycan motifs in the capsular polysaccharides of bacterial pathogens. These resources enable one to identify and study glycan motifs involved in immunogenicity, pathogenicity, molecular mimicry, and immune evasion, expanding our understanding of host-microbe interactions.
中文翻译:

用于研究糖介导的宿主-微生物相互作用的深度学习资源
聚糖是最多样化的生物聚合物,是由宿主微生物相互作用产生的进化压力形成的。在这里,我们介绍了机器学习和生物信息学方法,以利用聚糖中存在的进化信息来深入了解病原体和共生体如何与宿主相互作用。通过使用自然语言处理技术,我们开发了针对聚糖的深度学习模型,该模型在精选的19,299种独特聚糖数据集上进行了训练,可用于研究和预测聚糖功能。我们表明,这些模型可用于预测聚糖的免疫原性和细菌菌株的致病性,以及通过分子模拟研究聚糖介导的免疫逃逸。我们还开发了聚糖比对方法,并使用这些方法来分析细菌病原体荚膜多糖中确定毒力的聚糖基序。这些资源使人们能够识别和研究涉及免疫原性,致病性,分子模拟和免疫逃避的聚糖基序,从而扩大了我们对宿主-微生物相互作用的了解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号