Cell ( IF 45.5 ) Pub Date : 2020-10-24 , DOI: 10.1016/j.cell.2020.10.030 Zharko Daniloski 1 , Tristan X Jordan 2 , Hans-Hermann Wessels 1 , Daisy A Hoagland 2 , Silva Kasela 3 , Mateusz Legut 1 , Silas Maniatis 4 , Eleni P Mimitou 5 , Lu Lu 1 , Evan Geller 1 , Oded Danziger 2 , Brad R Rosenberg 2 , Hemali Phatnani 6 , Peter Smibert 5 , Tuuli Lappalainen 3 , Benjamin R tenOever 2 , Neville E Sanjana 1
|
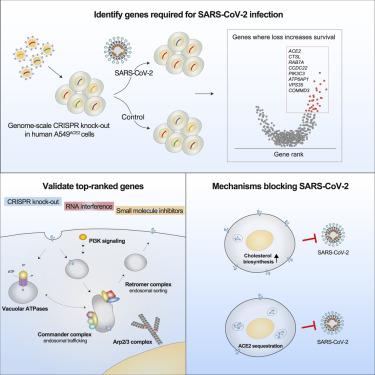
To better understand host-virus genetic dependencies and find potential therapeutic targets for COVID-19, we performed a genome-scale CRISPR loss-of-function screen to identify host factors required for SARS-CoV-2 viral infection of human alveolar epithelial cells. Top-ranked genes cluster into distinct pathways, including the vacuolar ATPase proton pump, Retromer, and Commander complexes. We validate these gene targets using several orthogonal methods such as CRISPR knockout, RNA interference knockdown, and small-molecule inhibitors. Using single-cell RNA-sequencing, we identify shared transcriptional changes in cholesterol biosynthesis upon loss of top-ranked genes. In addition, given the key role of the ACE2 receptor in the early stages of viral entry, we show that loss of RAB7A reduces viral entry by sequestering the ACE2 receptor inside cells. Overall, this work provides a genome-scale, quantitative resource of the impact of the loss of each host gene on fitness/response to viral infection.











































 京公网安备 11010802027423号
京公网安备 11010802027423号