当前位置:
X-MOL 学术
›
Biotechnol. Bioeng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A generalized machine‐learning aided method for targeted identification of industrial enzymes from metagenome: A xylanase temperature dependence case study
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2020-10-23 , DOI: 10.1002/bit.27608 Mehdi Foroozandeh Shahraki 1 , Kiana Farhadyar 1 , Kaveh Kavousi 1 , Mohammad H Azarabad 1 , Amin Boroomand 2 , Shohreh Ariaeenejad 3 , Ghasem Hosseini Salekdeh 3, 4
Biotechnology and Bioengineering ( IF 3.5 ) Pub Date : 2020-10-23 , DOI: 10.1002/bit.27608 Mehdi Foroozandeh Shahraki 1 , Kiana Farhadyar 1 , Kaveh Kavousi 1 , Mohammad H Azarabad 1 , Amin Boroomand 2 , Shohreh Ariaeenejad 3 , Ghasem Hosseini Salekdeh 3, 4
Affiliation
|
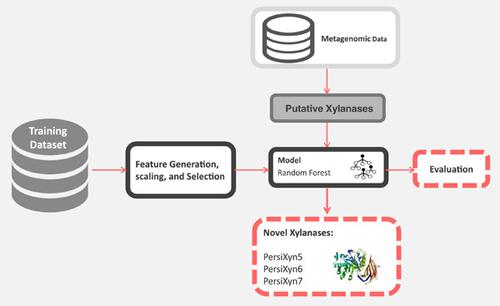
Growing industrial utilization of enzymes and the increasing availability of metagenomic data highlight the demand for effective methods of targeted identification and verification of novel enzymes from various environmental microbiota. Xylanases are a class of enzymes with numerous industrial applications and are involved in the degradation of xylose, a component of lignocellulose. The optimum temperature of enzymes is an essential factor to be considered when choosing appropriate biocatalysts for a particular purpose. Therefore, in silico prediction of this attribute is a significant cost and time‐effective step in the effort to characterize novel enzymes. The objective of this study was to develop a computational method to predict the thermal dependence of xylanases. This tool was then implemented for targeted screening of putative xylanases with specific thermal dependencies from metagenomic data and resulted in the identification of three novel xylanases from sheep and cow rumen microbiota. Here we present thermal activity prediction for xylanase, a new sequence‐based machine learning method that has been trained using a selected combination of various protein features. This random forest classifier discriminates non‐thermophilic, thermophilic, and hyper‐thermophilic xylanases. The model's performance was evaluated through multiple iterations of sixfold cross‐validations as well as holdout tests, and it is freely accessible as a web‐service at arimees.com.
中文翻译:

从宏基因组中靶向识别工业酶的通用机器学习辅助方法:木聚糖酶温度依赖性案例研究
酶的工业应用不断增长和宏基因组数据的可用性不断提高,突出了对来自各种环境微生物群的新型酶的靶向鉴定和验证的有效方法的需求。木聚糖酶是一类具有众多工业应用的酶,并参与木糖(木质纤维素的一种成分)的降解。在为特定目的选择合适的生物催化剂时,酶的最适温度是一个需要考虑的重要因素。因此,对该属性的计算机模拟预测是表征新型酶的重要成本和时间有效步骤。本研究的目的是开发一种计算方法来预测木聚糖酶的热依赖性。然后实施该工具以从宏基因组数据中靶向筛选具有特定热依赖性的推定木聚糖酶,并从羊和牛瘤胃微生物群中鉴定出三种新型木聚糖酶。在这里,我们提出了木聚糖酶的热活性预测,这是一种新的基于序列的机器学习方法,已使用各种蛋白质特征的选定组合进行训练。这种随机森林分类器区分非嗜热、嗜热和超嗜热木聚糖酶。该模型的性能通过六重交叉验证和坚持测试的多次迭代进行评估,并且它可以作为网络服务在 arimees.com 上免费访问。在这里,我们提出了木聚糖酶的热活性预测,这是一种新的基于序列的机器学习方法,已使用各种蛋白质特征的选定组合进行训练。这种随机森林分类器区分非嗜热、嗜热和超嗜热木聚糖酶。该模型的性能通过六重交叉验证和坚持测试的多次迭代进行评估,并且它可以作为网络服务在 arimees.com 上免费访问。在这里,我们提出了木聚糖酶的热活性预测,这是一种新的基于序列的机器学习方法,已使用各种蛋白质特征的选定组合进行训练。这种随机森林分类器区分非嗜热、嗜热和超嗜热木聚糖酶。该模型的性能通过六重交叉验证和坚持测试的多次迭代进行评估,并且它可以作为网络服务在 arimees.com 上免费访问。
更新日期:2020-10-23
中文翻译:

从宏基因组中靶向识别工业酶的通用机器学习辅助方法:木聚糖酶温度依赖性案例研究
酶的工业应用不断增长和宏基因组数据的可用性不断提高,突出了对来自各种环境微生物群的新型酶的靶向鉴定和验证的有效方法的需求。木聚糖酶是一类具有众多工业应用的酶,并参与木糖(木质纤维素的一种成分)的降解。在为特定目的选择合适的生物催化剂时,酶的最适温度是一个需要考虑的重要因素。因此,对该属性的计算机模拟预测是表征新型酶的重要成本和时间有效步骤。本研究的目的是开发一种计算方法来预测木聚糖酶的热依赖性。然后实施该工具以从宏基因组数据中靶向筛选具有特定热依赖性的推定木聚糖酶,并从羊和牛瘤胃微生物群中鉴定出三种新型木聚糖酶。在这里,我们提出了木聚糖酶的热活性预测,这是一种新的基于序列的机器学习方法,已使用各种蛋白质特征的选定组合进行训练。这种随机森林分类器区分非嗜热、嗜热和超嗜热木聚糖酶。该模型的性能通过六重交叉验证和坚持测试的多次迭代进行评估,并且它可以作为网络服务在 arimees.com 上免费访问。在这里,我们提出了木聚糖酶的热活性预测,这是一种新的基于序列的机器学习方法,已使用各种蛋白质特征的选定组合进行训练。这种随机森林分类器区分非嗜热、嗜热和超嗜热木聚糖酶。该模型的性能通过六重交叉验证和坚持测试的多次迭代进行评估,并且它可以作为网络服务在 arimees.com 上免费访问。在这里,我们提出了木聚糖酶的热活性预测,这是一种新的基于序列的机器学习方法,已使用各种蛋白质特征的选定组合进行训练。这种随机森林分类器区分非嗜热、嗜热和超嗜热木聚糖酶。该模型的性能通过六重交叉验证和坚持测试的多次迭代进行评估,并且它可以作为网络服务在 arimees.com 上免费访问。











































 京公网安备 11010802027423号
京公网安备 11010802027423号