当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
iGlu_AdaBoost: Identification of Lysine Glutarylation Using the AdaBoost Classifier
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-10-22 , DOI: 10.1021/acs.jproteome.0c00314 Lijun Dou 1, 2 , Xiaoling Li 3 , Lichao Zhang 4 , Huaikun Xiang 1 , Lei Xu 5
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2020-10-22 , DOI: 10.1021/acs.jproteome.0c00314 Lijun Dou 1, 2 , Xiaoling Li 3 , Lichao Zhang 4 , Huaikun Xiang 1 , Lei Xu 5
Affiliation
|
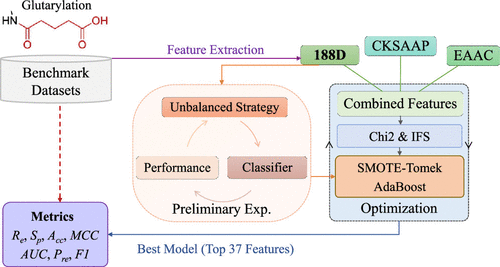
Lysine glutarylation is a newly reported post-translational modification (PTM) that plays significant roles in regulating metabolic and mitochondrial processes. Accurate identification of protein glutarylation is the primary task to better investigate molecular functions and various applications. Due to the common disadvantages of the time-consuming and expensive nature of traditional biological sequencing techniques as well as the explosive growth of protein data, building precise computational models to rapidly diagnose glutarylation is a popular and feasible solution. In this work, we proposed a novel AdaBoost-based predictor called iGlu_AdaBoost to distinguish glutarylation and non-glutarylation sequences. Here, the top 37 features were chosen from a total of 1768 combined features using Chi2 following incremental feature selection (IFS) to build the model, including 188D, the composition of k-spaced amino acid pairs (CKSAAP), and enhanced amino acid composition (EAAC). With the help of the hybrid-sampling method SMOTE-Tomek, the AdaBoost algorithm was performed with satisfactory recall, specificity, and AUC values of 87.48%, 72.49%, and 0.89 over 10-fold cross validation as well as 72.73%, 71.92%, and 0.63 over independent test, respectively. Further feature analysis inferred that positively charged amino acids RK play critical roles in glutarylation recognition. Our model presented the well generalization ability and consistency of the prediction results of positive and negative samples, which is comparable to four published tools. The proposed predictor is an efficient tool to find potential glutarylation sites and provides helpful suggestions for further research on glutarylation mechanisms and concerned disease treatments.
中文翻译:

iGlu_AdaBoost:使用AdaBoost分类器鉴定赖氨酸谷氨酸化
赖氨酸戊二酸化是新近报道的翻译后修饰(PTM),在调节代谢和线粒体过程中起重要作用。准确鉴定蛋白质的戊二酸化是更好地研究分子功能和各种应用的主要任务。由于传统生物测序技术耗时且昂贵的性质以及蛋白质数据的爆炸性增长的共同缺点,因此建立精确的计算模型以快速诊断戊二酸化是一种流行且可行的解决方案。在这项工作中,我们提出了一种新颖的基于AdaBoost的预测子,称为iGlu_AdaBoost,以区分戊二酸和非戊二酸序列。这里,ķ-间隔氨基酸对(CKSAAP)和增强的氨基酸组成(EAAC)。借助混合采样方法SMOTE-Tomek,AdaBoost算法执行时具有令人满意的召回率,特异性,并且经过10倍交叉验证的AUC值分别为87.48%,72.49%和0.89,以及72.73%,71.92%以及独立测试的0.63。进一步的特征分析推断带正电荷的氨基酸RK在戊二酰识别中起关键作用。我们的模型展示了正样本和负样本的预测结果的良好泛化能力和一致性,可与四个已发布的工具进行比较。所提出的预测因子是发现潜在的戊二酸位点的有效工具,并为进一步研究戊二酸化机理和相关疾病治疗提供了有益的建议。
更新日期:2020-10-22
中文翻译:

iGlu_AdaBoost:使用AdaBoost分类器鉴定赖氨酸谷氨酸化
赖氨酸戊二酸化是新近报道的翻译后修饰(PTM),在调节代谢和线粒体过程中起重要作用。准确鉴定蛋白质的戊二酸化是更好地研究分子功能和各种应用的主要任务。由于传统生物测序技术耗时且昂贵的性质以及蛋白质数据的爆炸性增长的共同缺点,因此建立精确的计算模型以快速诊断戊二酸化是一种流行且可行的解决方案。在这项工作中,我们提出了一种新颖的基于AdaBoost的预测子,称为iGlu_AdaBoost,以区分戊二酸和非戊二酸序列。这里,ķ-间隔氨基酸对(CKSAAP)和增强的氨基酸组成(EAAC)。借助混合采样方法SMOTE-Tomek,AdaBoost算法执行时具有令人满意的召回率,特异性,并且经过10倍交叉验证的AUC值分别为87.48%,72.49%和0.89,以及72.73%,71.92%以及独立测试的0.63。进一步的特征分析推断带正电荷的氨基酸RK在戊二酰识别中起关键作用。我们的模型展示了正样本和负样本的预测结果的良好泛化能力和一致性,可与四个已发布的工具进行比较。所提出的预测因子是发现潜在的戊二酸位点的有效工具,并为进一步研究戊二酸化机理和相关疾病治疗提供了有益的建议。











































 京公网安备 11010802027423号
京公网安备 11010802027423号