当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Bioinformatic Prediction of Gene Ontology Terms of Uncharacterized Proteins from Chromosome 11
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2020-10-22 , DOI: 10.1021/acs.jproteome.0c00482 Heeyoun Hwang 1 , Ji Eun Im 2 , Yeji Yang 1 , Hyejin Kim 1, 3 , Kyung-Hoon Kwon 1 , Yun-Hee Kim 2, 4 , Jin Young Kim 1 , Jong Shin Yoo 1, 3
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2020-10-22 , DOI: 10.1021/acs.jproteome.0c00482 Heeyoun Hwang 1 , Ji Eun Im 2 , Yeji Yang 1 , Hyejin Kim 1, 3 , Kyung-Hoon Kwon 1 , Yun-Hee Kim 2, 4 , Jin Young Kim 1 , Jong Shin Yoo 1, 3
Affiliation
|
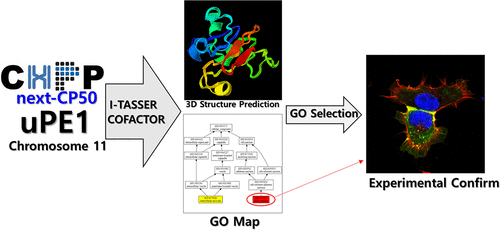
In chromosome 11, 71 out of its 1254 proteins remain functionally uncharacterized on the basis of their existence evidence (uPE1s) following the latest version of neXtProt (release 2020-01-17). Because in vivo and in vitro experimental strategies are often time-consuming and labor-intensive, there is a need for a bioinformatics tool to predict the function annotation. Here, we used I-TASSER/COFACTOR provided on the neXtProt web site, which predicts gene ontology (GO) terms based on the 3D structure of the protein. I-TASSER/COFACTOR predicted 2413 GO terms with a benchmark dataset of the 22 proteins belonging to PE1 of chromosome 11. In this study, we developed a filtering algorithm in order to select specific GO terms using the GO map generated by I-TASSER/COFACTOR. As a result, 187 specific GO terms showed a higher average precision-recall score at the least cellular component term compared to 2413 predicted GO terms. Next, we applied 65 proteins belonging to uPE1s of chromosome 11, and then 409 out of 6684 GO terms survived, where 103 and 142 GO terms of molecular function and biological process, respectively, were included. Representatively, the cellular component GO terms of CCDC90B, C11orf52, and the SMAP were predicted and validated using the overexpression system into 293T cells and immunofluorescence staining. We will further study their biological and molecular functions toward the goal of the neXt-CP50 project as a part of C-HPP. We shared all results and programs in Github (https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git).
中文翻译:

11号染色体未表征蛋白的基因本体学术语的生物信息学预测
在11号染色体上,根据最新版本的neXtProt(版本2020-01-17)的存在证据(uPE1s),其1254种蛋白质中的71种蛋白质仍未发挥功能。因为体内和体外实验策略通常是耗时且费力的,因此需要一种生物信息学工具来预测功能注释。在这里,我们使用了neXtProt网站上提供的I-TASSER / COFACTOR,它基于蛋白质的3D结构来预测基因本体论(GO)术语。I-TASSER / COFACTOR用属于11号染色体PE1的22种蛋白质的基准数据集预测了2413个GO项。在这项研究中,我们开发了一种过滤算法,以便使用由I-TASSER /生成的GO图来选择特定的GO项系数。结果,与2413个预测的GO项相比,在最少的细胞成分项中,有187个特定的GO项显示出更高的平均精确回忆得分。接下来,我们应用了65个属于11号染色体uPE1的蛋白质,然后在6684个GO项中有409个存活下来,其中分别包括分子功能和生物学过程的103和142个GO术语。代表性地,使用过表达系统进入293T细胞并进行免疫荧光染色,可以预测和验证CCDC90B,C11orf52和SMAP的细胞成分GO术语。我们将进一步研究它们的生物学和分子功能,以实现作为C-HPP一部分的neXt-CP50项目的目标。我们在Github(https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git)中共享了所有结果和程序。我们将进一步研究它们的生物学和分子功能,以实现作为C-HPP一部分的neXt-CP50项目的目标。我们在Github(https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git)中共享了所有结果和程序。我们将进一步研究其生物学和分子功能,以实现作为C-HPP一部分的neXt-CP50项目的目标。我们在Github(https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git)中共享了所有结果和程序。
更新日期:2020-12-04
中文翻译:

11号染色体未表征蛋白的基因本体学术语的生物信息学预测
在11号染色体上,根据最新版本的neXtProt(版本2020-01-17)的存在证据(uPE1s),其1254种蛋白质中的71种蛋白质仍未发挥功能。因为体内和体外实验策略通常是耗时且费力的,因此需要一种生物信息学工具来预测功能注释。在这里,我们使用了neXtProt网站上提供的I-TASSER / COFACTOR,它基于蛋白质的3D结构来预测基因本体论(GO)术语。I-TASSER / COFACTOR用属于11号染色体PE1的22种蛋白质的基准数据集预测了2413个GO项。在这项研究中,我们开发了一种过滤算法,以便使用由I-TASSER /生成的GO图来选择特定的GO项系数。结果,与2413个预测的GO项相比,在最少的细胞成分项中,有187个特定的GO项显示出更高的平均精确回忆得分。接下来,我们应用了65个属于11号染色体uPE1的蛋白质,然后在6684个GO项中有409个存活下来,其中分别包括分子功能和生物学过程的103和142个GO术语。代表性地,使用过表达系统进入293T细胞并进行免疫荧光染色,可以预测和验证CCDC90B,C11orf52和SMAP的细胞成分GO术语。我们将进一步研究它们的生物学和分子功能,以实现作为C-HPP一部分的neXt-CP50项目的目标。我们在Github(https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git)中共享了所有结果和程序。我们将进一步研究它们的生物学和分子功能,以实现作为C-HPP一部分的neXt-CP50项目的目标。我们在Github(https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git)中共享了所有结果和程序。我们将进一步研究其生物学和分子功能,以实现作为C-HPP一部分的neXt-CP50项目的目标。我们在Github(https://github.com/heeyounh/I-TASSER-COFACTOR-filtering.git)中共享了所有结果和程序。



























 京公网安备 11010802027423号
京公网安备 11010802027423号