当前位置:
X-MOL 学术
›
J. Zool. Syst. Evol. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Chromosomal analysis of eight species of dragonflies (Anisoptera) and damselflies (Zygoptera) using conventional cytogenetics and fluorescence in situ hybridization: Insights into the karyotype evolution of the ancient insect order Odonata
Journal of Zoological Systematics and Evolutionary Research ( IF 2.0 ) Pub Date : 2020-10-21 , DOI: 10.1111/jzs.12429 Valentina Kuznetsova 1 , Anna Maryańska‐Nadachowska 2 , Boris Anokhin 1 , Nazar Shapoval 1 , Anatoly Shapoval 3
Journal of Zoological Systematics and Evolutionary Research ( IF 2.0 ) Pub Date : 2020-10-21 , DOI: 10.1111/jzs.12429 Valentina Kuznetsova 1 , Anna Maryańska‐Nadachowska 2 , Boris Anokhin 1 , Nazar Shapoval 1 , Anatoly Shapoval 3
Affiliation
|
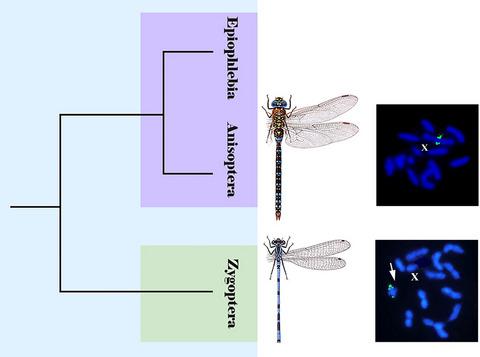
All Odonata species studied to date using fluorescence in situ hybridization (FISH) belong to the dragonfly (Anisoptera) families Corduliidae and Libellulidae. It was shown that 18S rRNA gene loci locate on one of the largest pairs of autosomes in every species, whereas the “insect” telomere motif (TTAGG)n is absent in all but one species. For better understanding the chromosomal organization and evolution of Odonata, we used C‐banding and FISH to study the karyotypes and map TTAGG sequences and major rRNA loci on chromosomes of three more dragonfly species from the families Corduliidae, Libellulidae, and Aeshnidae. Moreover, we obtained the first FISH‐data on the suborder Zygoptera (damselflies) by analyzing five species of the families Coenagrionidae and Calopterygidae. We showed that all studied dragonfly species had 2n = 24A + X. The same karyotype was observed in the damselfly family Coenagrionidae, whereas in species of the Calopterygidae, the karyotype 2n = 26A + X was found. Both dragonfly and damselfly species had a pair of m‐chromosomes; constitutive heterochromatin tended to be concentrated in the terminal regions of their chromosomes. The use of (TTAGG)n and 18S rRNA gene probes in dual‐color FISH did not generate (TTAGG)n fluorescent signals in any species; major rRNA clusters were revealed on one of the largest pairs of autosomes in all Anisoptera species but on m‐chromosomes in all Zygoptera species. Our results suggest that the former 18S location pattern was ancestral in the Odonata and the latter pattern had an ancient origin and could arise in a common ancestor of the damselfly superfamilies Calopterygoidea and Coenagrionoidea.
中文翻译:

使用常规细胞遗传学和荧光原位杂交技术对8种蜻蜓(Anisoptera)和豆娘(Zygoptera)进行染色体分析:洞悉古代昆虫纲Odonata的核型进化
迄今为止,所有使用荧光原位杂交技术(FISH)研究的Odonata物种都属于蜻蜓(Anisoptera)科Corduliidae和Libellulidae。结果表明,18S rRNA基因位点位于每个物种中最大的常染色体对之一上,而“昆虫”端粒基序(TTAGG)n除了一个物种外,其他所有物种都不存在。为了更好地了解Odonata的染色体组织和进化,我们使用C带和FISH研究了核型,并在来自Corduliidae,Libellulidae和Aeshnidae的另外三种蜻蜓物种的染色体上绘制了TTAGG序列和主要rRNA基因座。此外,通过分析Coenagrionidae科和Calopterygidae科的5种物种,我们获得了Zygoptera(豆娘)亚目的第一个FISH数据。我们表明,所有研究的蜻蜓物种都具有2 n = 24A +X。在豆娘科Coenagrionidae中观察到了相同的核型,而在Calopterygidae物种中,核型为2 n。 = 26A +X。蜻蜓和豆娘物种都有一对间染色体。组成型异染色质倾向于集中在其染色体的末端区域。在双色FISH中使用(TTAGG)n和18S rRNA基因探针不会在任何物种中产生(TTAGG)n荧光信号。在所有Anisoptera物种中最大的常染色体对之一上揭示了主要的rRNA簇,但在所有Zygoptera物种中的m染色体上揭示了主要rRNA簇。我们的研究结果表明,前18S的定位模式在Odonata中是祖先的,而后一种模式则起源于古老,可能起源于豆娘超家族Calopterygoidea和Coenagrionoidea的共同祖先。
更新日期:2020-10-21
中文翻译:

使用常规细胞遗传学和荧光原位杂交技术对8种蜻蜓(Anisoptera)和豆娘(Zygoptera)进行染色体分析:洞悉古代昆虫纲Odonata的核型进化
迄今为止,所有使用荧光原位杂交技术(FISH)研究的Odonata物种都属于蜻蜓(Anisoptera)科Corduliidae和Libellulidae。结果表明,18S rRNA基因位点位于每个物种中最大的常染色体对之一上,而“昆虫”端粒基序(TTAGG)n除了一个物种外,其他所有物种都不存在。为了更好地了解Odonata的染色体组织和进化,我们使用C带和FISH研究了核型,并在来自Corduliidae,Libellulidae和Aeshnidae的另外三种蜻蜓物种的染色体上绘制了TTAGG序列和主要rRNA基因座。此外,通过分析Coenagrionidae科和Calopterygidae科的5种物种,我们获得了Zygoptera(豆娘)亚目的第一个FISH数据。我们表明,所有研究的蜻蜓物种都具有2 n = 24A +X。在豆娘科Coenagrionidae中观察到了相同的核型,而在Calopterygidae物种中,核型为2 n。 = 26A +X。蜻蜓和豆娘物种都有一对间染色体。组成型异染色质倾向于集中在其染色体的末端区域。在双色FISH中使用(TTAGG)n和18S rRNA基因探针不会在任何物种中产生(TTAGG)n荧光信号。在所有Anisoptera物种中最大的常染色体对之一上揭示了主要的rRNA簇,但在所有Zygoptera物种中的m染色体上揭示了主要rRNA簇。我们的研究结果表明,前18S的定位模式在Odonata中是祖先的,而后一种模式则起源于古老,可能起源于豆娘超家族Calopterygoidea和Coenagrionoidea的共同祖先。











































 京公网安备 11010802027423号
京公网安备 11010802027423号