Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2020-10-20 , DOI: 10.1016/j.jbi.2020.103605 Chunjiang Yu 1 , Xin Qi 2 , Yuxin Lin 3 , Yin Li 3 , Bairong Shen 4
|
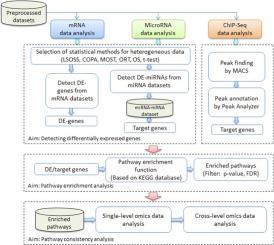
The latest advances in the next generation sequencing technology have greatly facilitated the extensive research of genomics and transcriptomics, thereby promoting the decoding of carcinogenesis with unprecedented resolution. Considering the contribution of analyzing high-throughput multi-omics data to the exploration of cancer molecular mechanisms, an integrated tool for heterogeneous multi-omics data analysis (iODA) is proposed for the systems-level interpretation of multi-omics data, i.e., transcriptomic profiles (mRNA or miRNA expression data) and protein-DNA interactions (ChIP-Seq data). Considering the data heterogeneity, iODA can compare six statistical algorithms in differential analysis for the selected sample data and assist users in choosing the globally optimal one for dysfunctional mRNA or miRNA identification. Since molecular signatures are more consistent at the pathway level than at the gene level, the tool is able to enrich the identified dysfunctional molecules onto the KEGG pathways and extracted the consistent items as key components for further pathogenesis investigation. Compared with other tools, iODA is multi-functional for the systematic analysis of different level of omics data, and its analytical power was demonstrated through case studies of single and cross-level prostate cancer omics data. iODA is open source under GNU GPL and can be downloaded from http://www.sysbio.org.cn/iODA.
中文翻译:

iODA:从异质多组学数据分析癌症途径一致性的综合工具
下一代测序技术的最新进展极大地促进了基因组学和转录组学的广泛研究,从而以前所未有的分辨率促进了癌变的解码。考虑到分析高通量多组学数据对探索癌症分子机制的贡献,提出了一种用于异类多组学数据分析(iODA)的集成工具,用于系统级解释多组学数据,即转录组学概况(mRNA或miRNA表达数据)和蛋白质-DNA相互作用(ChIP-Seq数据)。考虑到数据的异质性,iODA可以在差异分析中比较六种统计算法,以用于选定的样本数据,并帮助用户选择功能失调的mRNA或miRNA识别的全局最佳算法。由于在路径水平上比在基因水平上分子标记更加一致,该工具能够将识别出的功能障碍分子富集到KEGG途径上,并提取出一致的项目作为进一步发病机理研究的关键组成部分。与其他工具相比,iODA具有多功能性,可以对不同水平的组学数据进行系统分析,并且通过对单级和跨级前列腺癌组学数据的案例研究证明了其分析能力。iODA是GNU GPL下的开放源代码,可以从http://www.sysbio.org.cn/iODA下载。与其他工具相比,iODA具有多功能性,可以对不同水平的组学数据进行系统分析,并且通过对单级和跨级前列腺癌组学数据的案例研究证明了其分析能力。iODA是GNU GPL下的开源软件,可以从http://www.sysbio.org.cn/iODA下载。与其他工具相比,iODA具有多功能性,可以对不同水平的组学数据进行系统分析,并且通过对单级和跨级前列腺癌组学数据的案例研究证明了其分析能力。iODA是GNU GPL下的开源软件,可以从http://www.sysbio.org.cn/iODA下载。











































 京公网安备 11010802027423号
京公网安备 11010802027423号