当前位置:
X-MOL 学术
›
J. Leukoc. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Single‐cell transcriptomics uncover distinct innate and adaptive cell subsets during tissue homeostasis and regeneration
Journal of Leukocyte Biology ( IF 3.6 ) Pub Date : 2020-10-18 , DOI: 10.1002/jlb.6mr0720-131r Kevin Y. Yang 1 , Manching Ku 2 , Kathy O. Lui 1, 3
Journal of Leukocyte Biology ( IF 3.6 ) Pub Date : 2020-10-18 , DOI: 10.1002/jlb.6mr0720-131r Kevin Y. Yang 1 , Manching Ku 2 , Kathy O. Lui 1, 3
Affiliation
|
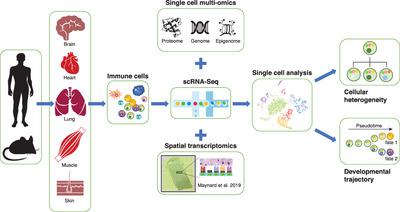
Recently, immune cell‐mediated tissue repair and regeneration has been an emerging paradigm of regenerative medicine. Immune cells form an essential part of the wound as induction of inflammation is a necessary step to elicit tissue healing. Rapid progress in transcriptomic analyses by high‐throughput next‐generation sequencing has been developed to study gene regulatory network and establish molecular signatures of immune cells that could potentially predict their functional roles in tissue repair and regeneration. However, the identification of cellular heterogeneity especially on the rare cell subsets has been limited in transcriptomic analyses of bulk cell populations. Therefore, genome‐wide, single‐cell RNA sequencing (scRNA‐Seq) has offered an unprecedented approach to unravel cellular diversity and to study novel immune cell populations involved in tissue repair and regeneration through unsupervised sampling of individual cells without the need to rely on prior knowledge about cell‐specific markers. The analysis of gene expression patterns at a single‐cell resolution also holds promises to uncover the mechanisms and therefore the development of therapeutic strategy promoting immunoregenerative medicine. In this review, we will discuss how scRNA‐Seq facilitates the characterization of immune cells, including macrophages, innate lymphoid cells and T and B lymphocytes, discovery of immune cell heterogeneity, identification of novel subsets, and tracking of developmental trajectories of distinct immune cells during tissue homeostasis, repair, and regeneration.
中文翻译:

单细胞转录组学在组织稳态和再生过程中揭示了不同的先天和适应性细胞亚群
最近,免疫细胞介导的组织修复和再生已成为再生医学的新兴范例。免疫细胞形成伤口的重要组成部分,因为炎症的诱导是引发组织愈合的必要步骤。已经开发出通过高通量下一代测序技术进行转录组学分析的快速进展,以研究基因调控网络并建立免疫细胞的分子特征,从而可能预测其在组织修复和再生中的功能。然而,细胞异质性的鉴定,特别是在稀有细胞亚群上的鉴定,在大细胞群体的转录组分析中受到了限制。因此,全基因组 单细胞RNA测序(scRNA‐Seq)提供了一种前所未有的方法来揭示细胞多样性,并通过无监督地采样单个细胞来研究参与组织修复和再生的新型免疫细胞群体,而无需依赖于有关细胞特异性的现有知识标记。以单细胞分辨率对基因表达模式进行分析也有望揭示其机制,从而开发促进免疫再生医学的治疗策略。在本文中,我们将讨论scRNA‐Seq如何促进免疫细胞的表征,包括巨噬细胞,先天淋巴样细胞以及T和B淋巴细胞,发现免疫细胞异质性,鉴定新的亚群以及跟踪不同免疫细胞的发育轨迹组织动态平衡期间
更新日期:2020-10-30
中文翻译:

单细胞转录组学在组织稳态和再生过程中揭示了不同的先天和适应性细胞亚群
最近,免疫细胞介导的组织修复和再生已成为再生医学的新兴范例。免疫细胞形成伤口的重要组成部分,因为炎症的诱导是引发组织愈合的必要步骤。已经开发出通过高通量下一代测序技术进行转录组学分析的快速进展,以研究基因调控网络并建立免疫细胞的分子特征,从而可能预测其在组织修复和再生中的功能。然而,细胞异质性的鉴定,特别是在稀有细胞亚群上的鉴定,在大细胞群体的转录组分析中受到了限制。因此,全基因组 单细胞RNA测序(scRNA‐Seq)提供了一种前所未有的方法来揭示细胞多样性,并通过无监督地采样单个细胞来研究参与组织修复和再生的新型免疫细胞群体,而无需依赖于有关细胞特异性的现有知识标记。以单细胞分辨率对基因表达模式进行分析也有望揭示其机制,从而开发促进免疫再生医学的治疗策略。在本文中,我们将讨论scRNA‐Seq如何促进免疫细胞的表征,包括巨噬细胞,先天淋巴样细胞以及T和B淋巴细胞,发现免疫细胞异质性,鉴定新的亚群以及跟踪不同免疫细胞的发育轨迹组织动态平衡期间










































 京公网安备 11010802027423号
京公网安备 11010802027423号