当前位置:
X-MOL 学术
›
FEBS Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
How chromosome topologies get their shape: views from proximity ligation and microscopy methods
FEBS Letters ( IF 3.0 ) Pub Date : 2020-11-01 , DOI: 10.1002/1873-3468.13961 Yike Huang 1 , Roel Neijts 1 , Wouter Laat 1
FEBS Letters ( IF 3.0 ) Pub Date : 2020-11-01 , DOI: 10.1002/1873-3468.13961 Yike Huang 1 , Roel Neijts 1 , Wouter Laat 1
Affiliation
|
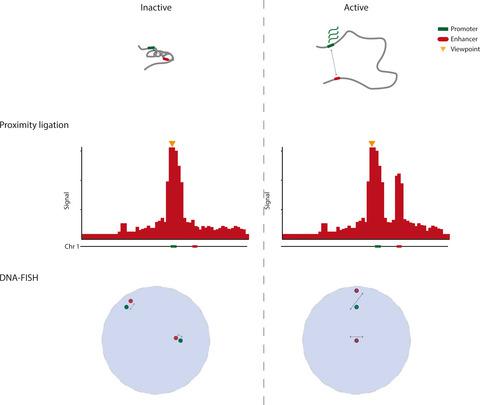
The 3D organization of our genome is an important determinant for the transcriptional output of a gene in (patho)physiological contexts. The spatial organization of linear chromosomes within nucleus is dominantly inferred using two distinct approaches, chromosome conformation capture (3C) and DNA fluorescent in situ hybridization (DNA–FISH). While 3C and its derivatives score genomic interaction frequencies based on proximity ligation events, DNA–FISH methods measure physical distances between genomic loci. Despite these approaches probe different characteristics of chromosomal topologies, they provide a coherent picture of how chromosomes are organized in higher‐order structures encompassing chromosome territories, compartments, and topologically associating domains. Yet, at the finer topological level of promoter–enhancer communication, the imaging‐centered and the 3C methods give more divergent and sometimes seemingly paradoxical results. Here, we compare and contrast observations made applying visual DNA–FISH and molecular 3C approaches. We emphasize that the 3C approach, due to its inherently competitive ligation step, measures only ‘relative’ proximities. A 3C interaction enriched between loci, therefore does not necessarily translates into a decrease in absolute spatial distance. Hence, we advocate caution when modeling chromosome conformations.
中文翻译:

染色体拓扑如何形成它们的形状:来自邻近连接和显微镜方法的观点
我们基因组的 3D 组织是(病理)生理环境中基因转录输出的重要决定因素。细胞核内线性染色体的空间组织主要使用两种不同的方法推断,染色体构象捕获 (3C) 和 DNA 荧光原位杂交 (DNA-FISH)。3C 及其衍生物根据邻近连接事件对基因组相互作用频率进行评分,而 DNA-FISH 方法则测量基因组位点之间的物理距离。尽管这些方法探测了染色体拓扑的不同特征,但它们提供了有关染色体如何在包含染色体区域、区室和拓扑关联域的高阶结构中组织的连贯图片。然而,在启动子-增强子通信的更精细拓扑水平上,以成像为中心和 3C 方法给出了更多不同的结果,有时似乎是矛盾的结果。在这里,我们比较和对比了应用视觉 DNA-FISH 和分子 3C 方法进行的观察。我们强调 3C 方法,由于其固有的竞争性结扎步骤,仅测量“相对”接近度。因此,基因座之间丰富的 3C 相互作用并不一定会转化为绝对空间距离的减少。因此,我们提倡在对染色体构象建模时要谨慎。因此,基因座之间丰富的 3C 相互作用并不一定会转化为绝对空间距离的减少。因此,我们提倡在对染色体构象建模时要谨慎。因此,基因座之间丰富的 3C 相互作用并不一定会转化为绝对空间距离的减少。因此,我们提倡在对染色体构象建模时要谨慎。
更新日期:2020-11-01
中文翻译:

染色体拓扑如何形成它们的形状:来自邻近连接和显微镜方法的观点
我们基因组的 3D 组织是(病理)生理环境中基因转录输出的重要决定因素。细胞核内线性染色体的空间组织主要使用两种不同的方法推断,染色体构象捕获 (3C) 和 DNA 荧光原位杂交 (DNA-FISH)。3C 及其衍生物根据邻近连接事件对基因组相互作用频率进行评分,而 DNA-FISH 方法则测量基因组位点之间的物理距离。尽管这些方法探测了染色体拓扑的不同特征,但它们提供了有关染色体如何在包含染色体区域、区室和拓扑关联域的高阶结构中组织的连贯图片。然而,在启动子-增强子通信的更精细拓扑水平上,以成像为中心和 3C 方法给出了更多不同的结果,有时似乎是矛盾的结果。在这里,我们比较和对比了应用视觉 DNA-FISH 和分子 3C 方法进行的观察。我们强调 3C 方法,由于其固有的竞争性结扎步骤,仅测量“相对”接近度。因此,基因座之间丰富的 3C 相互作用并不一定会转化为绝对空间距离的减少。因此,我们提倡在对染色体构象建模时要谨慎。因此,基因座之间丰富的 3C 相互作用并不一定会转化为绝对空间距离的减少。因此,我们提倡在对染色体构象建模时要谨慎。因此,基因座之间丰富的 3C 相互作用并不一定会转化为绝对空间距离的减少。因此,我们提倡在对染色体构象建模时要谨慎。











































 京公网安备 11010802027423号
京公网安备 11010802027423号