Gene ( IF 2.6 ) Pub Date : 2020-10-15 , DOI: 10.1016/j.gene.2020.145216 L. Sadeghi-Amiri , A. Barzegar , N. Nikbakhsh-Zati , P. Mehraban
|
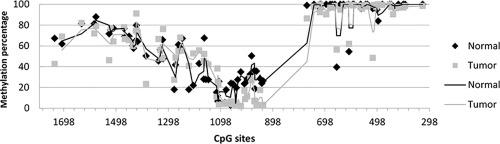
Owing to its broad substrate specificity of mainly xenobiotic and its preferential extrahepatic expression, cytochrome P450 1A1 (CYP1A1) is a principle member of the CYP450 detoxifying enzyme superfamily involving in carcinogenesis. In this study, we analyzed methylation status of 93 CpG sites, densely scattered within approximately 1.5 kb 5’ regulatory region of CYP1A1, and its association with gene transcription in tissue cohorts dissected from 40 patients with gastric cancer. Bisulfite sequencing and the resulting methylation percentages revealed dynamically methylated CpG sites located within or around xenobiotic response elements (XRE) 4-10, and a region of consistent hypermethylation located near proximal promoter, encompassing XRE2-3. Statistical analysis revealed significant differences of the methylation percentages at CpG sites -1415 (0.032) and -1524 (P=0.041) (located at the close upstream region of XRE10) between cancerous and normal gastric tissues as well as between those with and without lymphatic involvement. Quantitative real time PCR analysis showed that the CYP1A1 gene expression significantly increased in cancerous tissues compared to their normal tissue cohort, and is significantly associated with hypomethylation at the CpG sites -1383 (P=0.018) existing within the XRE10 motif. These data suggest that the variably methylated CpG site existing in the 5’ regulatory region of CYP1A1 gene, corresponding with the XRE10 regulatory region, is associated with the CYP1A1 gene upregulation and likely involved in gastric cancer incidence and metastasis. Methylation analysis of the CpG sites located within or around the XRE10 motif of the CYP1A1 promoter can be used as a potential marker to evaluate individual susceptibility to gastric cancer
中文翻译:

XRE -1383位的亚甲基化与胃腺癌中CYP1A1的上调有关
由于其主要是异种生物的广泛底物特异性和其优先的肝外表达,细胞色素P450 1A1(CYP1A1)是CYP450排毒酶超家族的主要成员,参与了致癌作用。在这项研究中,我们分析了93个CpG位点的甲基化状态,这些位点密集地分布在CYP1A1的约1.5 kb 5'调节区内,并与40例胃癌患者的组织队列中的基因转录相关。亚硫酸氢盐测序和所得的甲基化百分比揭示了位于异源生物反应元件(XRE)4-10内或周围的动态甲基化CpG位点,以及位于近端启动子附近的一致的高甲基化区域,包括XRE2-3。统计分析表明,癌组织和正常胃组织以及有无淋巴组织的CpG位点-1415(0.032)和-1524(P = 0.041)(位于XRE10的上游附近)的甲基化百分比存在显着差异参与。实时定量PCR分析表明CYP1A1与正常组织队列相比,该基因在癌组织中的表达明显增加,并且与XRE10基序中存在的CpG位点-1383(P = 0.018)的甲基化程度显着相关。这些数据表明存在于CYP1A1基因5'调节区中的甲基化CpG位点,与XRE10调节区相对应,与CYP1A1基因上调有关,并可能参与胃癌的发生和转移。位于CYP1A1启动子的XRE10基序内或周围的CpG位点的甲基化分析可用作评估个体对胃癌易感性的潜在标记











































 京公网安备 11010802027423号
京公网安备 11010802027423号