Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-10-14 , DOI: 10.1016/j.csbj.2020.10.010 Sebastien Leblanc 1, 2 , Marie A Brunet 1, 2
|
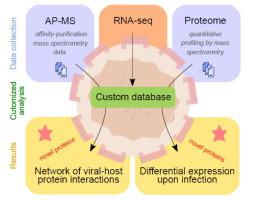
The Zika virus is a flavivirus that can cause fulminant outbreaks and lead to Guillain-Barré syndrome, microcephaly and fetal demise. Like other flaviviruses, the Zika virus is transmitted by mosquitoes and provokes neurological disorders. Despite its risk to public health, no antiviral nor vaccine are currently available. In the recent years, several studies have set to identify human host proteins interacting with Zika viral proteins to better understand its pathogenicity. Yet these studies used standard human protein sequence databases. Such databases rely on genome annotations, which enforce a minimal open reading frame (ORF) length criterion. An ever-increasing number of studies have demonstrated the shortcomings of such annotation, which overlooks thousands of functional ORFs. Here we show that the use of a customized database including currently non-annotated proteins led to the identification of 4 alternative proteins as interactors of the viral capsid and NS4A proteins. Furthermore, 12 alternative proteins were identified in the proteome profiling of Zika infected monocytes, one of which was significantly up-regulated. This study presents a computational framework for the re-analysis of proteomics datasets to better investigate the viral-host protein interplays upon infection with the Zika virus.
中文翻译:

使用更深入的 ORF 注释和转录组学对病原体-宿主系统进行建模,为蛋白质组学分析提供信息
寨卡病毒是一种黄病毒,可引起暴发性疫情,并导致格林-巴利综合征、小头畸形和胎儿死亡。与其他黄病毒一样,寨卡病毒通过蚊子传播并引发神经系统疾病。尽管对公共健康构成风险,但目前尚无抗病毒药物或疫苗。近年来,多项研究开始鉴定与寨卡病毒蛋白相互作用的人类宿主蛋白,以更好地了解其致病性。然而这些研究使用了标准的人类蛋白质序列数据库。此类数据库依赖于基因组注释,强制执行最小开放阅读框 (ORF) 长度标准。越来越多的研究已经证明了这种注释的缺点,它忽略了数千个功能性 ORF。在这里,我们展示了使用包含当前未注释蛋白质的定制数据库导致鉴定出 4 种替代蛋白质作为病毒衣壳和 NS4A 蛋白质的相互作用物。此外,在寨卡病毒感染的单核细胞的蛋白质组分析中发现了 12 种替代蛋白质,其中一种蛋白质显着上调。这项研究提出了一个用于重新分析蛋白质组数据集的计算框架,以更好地研究寨卡病毒感染后病毒与宿主蛋白质的相互作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号