Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2020-10-13 , DOI: 10.1016/j.jmb.2020.10.004 Witold Dyrka , Virginie Coustou , Asen Daskalov , Alons Lends , Thierry Bardin , Mélanie Berbon , Brice Kauffmann , Corinne Blancard , Bénédicte Salin , Antoine Loquet , Sven J. Saupe
|
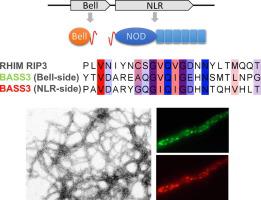
In filamentous fungi, amyloid signaling sequences allow Nod-like receptors (NLRs) to activate downstream cell-death inducing proteins with HeLo and HeLo-like (HELL) domains and amyloid RHIM and RHIM-related motifs control immune defense pathways in mammals and flies. Herein, we show bioinformatically that analogous amyloid signaling motifs exist in bacteria. These short motifs are found at the N terminus of NLRs and at the C terminus of proteins with a domain we term BELL. The corresponding NLR and BELL proteins are encoded by adjacent genes. We identify 10 families of such bacterial amyloid signaling sequences (BASS), one of which (BASS3) is homologous to RHIM and a fungal amyloid motif termed PP. BASS motifs occur nearly exclusively in bacteria forming multicellular structures (mainly in Actinobacteria and Cyanobacteria). We analyze experimentally a subset of seven of these motifs (from the most common BASS1 family and the RHIM-related BASS3 family) and find that these sequences form fibrils in vitro. Using a fungal in vivo model, we show that all tested BASS-motifs form prions and that the NLR-side motifs seed prion-formation of the corresponding BELL-side motif. We find that BASS3 motifs show partial prion cross-seeding with mammalian RHIM and fungal PP-motifs and that proline mutations on key positions of the BASS3 core motif, conserved in RHIM and PP-motifs, abolish prion formation. This work expands the paradigm of prion amyloid signaling to multicellular prokaryotes and suggests a long-term evolutionary conservation of these motifs from bacteria, to fungi and animals.
中文翻译:

细菌基因组中与NLR相关的淀粉样蛋白信号基元的鉴定
在丝状真菌中,淀粉样蛋白信号序列允许Nod样受体(NLR)激活具有HeLo和HeLo样(HELL)结构域的下游细胞死亡诱导蛋白,淀粉样蛋白RHIM和RHIM相关的基序控制哺乳动物和果蝇的免疫防御途径。在本文中,我们从生物学上证明了细菌中存在类似的淀粉样蛋白信号传导基序。这些短基序位于NLR的N末端和具有我们称为BELL的结构域的蛋白质的C末端。相应的NLR和BELL蛋白由相邻基因编码。我们确定这种细菌淀粉样蛋白信号序列(BASS)的10个家庭,其中之一(BASS3)与RHIM和称为PP的真菌淀粉样蛋白基序同源。BASS基序几乎只在形成多细胞结构的细菌中发生(主要发生在放线菌和蓝细菌)。我们通过实验分析了这些基序中的七个(来自最常见的BASS1家族和RHIM相关的BASS3家族)的子集,并发现这些序列在体外形成了原纤维。使用真菌体内模型,我们显示了所有测试的BASS-基序均形成病毒,而NLR侧基序则是相应BELL侧基序的pr病毒形成的种子。我们发现BASS3基序显示与哺乳动物RHIM和真菌PP-基序的部分病毒交叉播种,并且在RHIM和PP基序中保守的BASS3核心基序关键位置的脯氨酸突变消除了,病毒的形成。 这项工作将病毒淀粉样蛋白信号转导的范式扩展到多细胞原核生物,并提出了这些基序从细菌到真菌和动物的长期进化保守性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号