当前位置:
X-MOL 学术
›
Acta Cryst. D
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Automated orientation of water molecules in neutron crystallographic structures of proteins
Acta Crystallographica Section D ( IF 2.6 ) Pub Date : 2020-10-08 , DOI: 10.1107/s2059798320011729 Axl Eriksson 1 , Octav Caldararu 1 , Ulf Ryde 1 , Esko Oksanen 2
Acta Crystallographica Section D ( IF 2.6 ) Pub Date : 2020-10-08 , DOI: 10.1107/s2059798320011729 Axl Eriksson 1 , Octav Caldararu 1 , Ulf Ryde 1 , Esko Oksanen 2
Affiliation
|
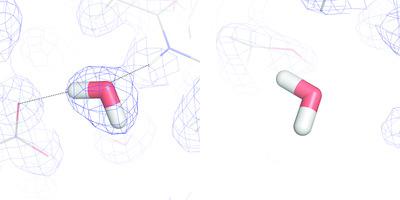
The structure and function of proteins are strongly affected by the surrounding solvent water, for example through hydrogen bonds and the hydrophobic effect. These interactions depend not only on the position, but also on the orientation, of the water molecules around the protein. Therefore, it is often vital to know the detailed orientations of the surrounding ordered water molecules. Such information can be obtained by neutron crystallography. However, it is tedious and time‐consuming to determine the correct orientation of every water molecule in a structure (there are typically several hundred of them), which is presently performed by manual evaluation. Here, a method has been developed that reliably automates the orientation of a water molecules in a simple and relatively fast way. Firstly, a quantitative quality measure, the real‐space correlation coefficient, was selected, together with a threshold that allows the identification of water molecules that are oriented. Secondly, the refinement procedure was optimized by varying the refinement method and parameters, thus finding settings that yielded the best results in terms of time and performance. It turned out to be favourable to employ only the neutron data and a fixed protein structure when reorienting the water molecules. Thirdly, a method has been developed that identifies and reorients inadequately oriented water molecules systematically and automatically. The method has been tested on three proteins, galectin‐3C, rubredoxin and inorganic pyrophosphatase, and it is shown that it yields improved orientations of the water molecules for all three proteins in a shorter time than manual model building. It also led to an increased number of hydrogen bonds involving water molecules for all proteins.
中文翻译:

蛋白质中子晶体结构中水分子的自动取向
蛋白质的结构和功能会受到周围溶剂水的强烈影响,例如通过氢键和疏水作用。这些相互作用不仅取决于蛋白质周围水分子的位置,还取决于其方向。因此,了解周围有序水分子的详细方向通常至关重要。这样的信息可以通过中子晶体学获得。但是,确定结构中每个水分子的正确方向(通常有数百个)的正确方向既繁琐又费时,目前这是通过人工评估来完成的。在此,已经开发出一种以简单且相对快速的方式可靠地使水分子的取向自动化的方法。首先,定量质量衡量 选择了真实空间相关系数,以及一个阈值,该阈值允许识别定向的水分子。其次,通过更改优化方法和参数来优化优化过程,从而找到在时间和性能方面产生最佳结果的设置。事实证明,在重新定向水分子时仅采用中子数据和固定的蛋白质结构是有利的。第三,已经开发出一种方法,该方法可以系统地,自动地识别和重新定向不充分的水分子。该方法已在三种蛋白质(半乳凝素3C,红氧化还原酶和无机焦磷酸酶)上进行了测试,结果表明,与手动建模相比,该方法可在较短的时间内对所有三种蛋白质产生改善的水分子方向。
更新日期:2020-10-08
中文翻译:

蛋白质中子晶体结构中水分子的自动取向
蛋白质的结构和功能会受到周围溶剂水的强烈影响,例如通过氢键和疏水作用。这些相互作用不仅取决于蛋白质周围水分子的位置,还取决于其方向。因此,了解周围有序水分子的详细方向通常至关重要。这样的信息可以通过中子晶体学获得。但是,确定结构中每个水分子的正确方向(通常有数百个)的正确方向既繁琐又费时,目前这是通过人工评估来完成的。在此,已经开发出一种以简单且相对快速的方式可靠地使水分子的取向自动化的方法。首先,定量质量衡量 选择了真实空间相关系数,以及一个阈值,该阈值允许识别定向的水分子。其次,通过更改优化方法和参数来优化优化过程,从而找到在时间和性能方面产生最佳结果的设置。事实证明,在重新定向水分子时仅采用中子数据和固定的蛋白质结构是有利的。第三,已经开发出一种方法,该方法可以系统地,自动地识别和重新定向不充分的水分子。该方法已在三种蛋白质(半乳凝素3C,红氧化还原酶和无机焦磷酸酶)上进行了测试,结果表明,与手动建模相比,该方法可在较短的时间内对所有三种蛋白质产生改善的水分子方向。











































 京公网安备 11010802027423号
京公网安备 11010802027423号