Journal of Molecular Graphics and Modelling ( IF 2.9 ) Pub Date : 2020-10-08 , DOI: 10.1016/j.jmgm.2020.107774 Md Abdullah-Al-Kamran Khan 1 , Rafeed Rahman Turjya 1 , Abul Bashar Mir Md Khademul Islam 1
|
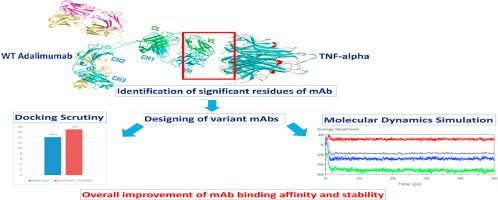
Amongst the anti-TNF-α therapy for rheumatoid arthritis and other autoimmune diseases, Adalimumab mAb is one of the best candidates. However, several risk factors are found to be associated with higher doses. Improvement of the binding properties will therefore significantly increase its therapeutic efficacy, reduce the dosage requirements, and ultimately the associated toxicity and treatment cost. Here, we proposed a systematic in silico approach of finding newer mAb variants with improved binding properties. Using various bioinformatics tools, we have identified the significant amino acid residues on Adalimumab mAb. Next, we searched for the suitability of the other residues for mutating the significant residues and from the combinations of suitable mutations, variants were designed. To find the most significant ones, binding properties of the variants were compared with the wild type Adalimumab mAb using molecular docking scrutiny and molecular dynamics simulation. Finally, structural properties between the variant and wild type were analyzed. We have identified the six most significant residues on Adalimumab mAb involved in the antigen-antibody interactions. Using the suitable mutations replacing each of these residues, we have modeled 143 variants. From several docking analyses, we have found five significant variants and after molecular dynamics simulation, one most significant variant with improved binding affinity was identified whose structural properties are similar to the wild type Adalimumab mAb. Designed variant from this study, may provide newer insights on the structure-based affinity improvements of monoclonal antibodies and likewise modifications of the Fc region will also improve the therapeutic effector functions of antibodies too.
中文翻译:

计算工程阿达木单抗单克隆抗体的结合亲和力,以设计潜在的生物仿制药
在针对类风湿关节炎和其他自身免疫性疾病的抗TNF-α治疗方法中,阿达木单抗mAb是最佳候选药物之一。但是,发现一些危险因素与较高剂量有关。因此,结合性能的改善将显着提高其治疗功效,降低剂量需求,并最终降低相关的毒性和治疗成本。在这里,我们提出了系统的计算机模拟寻找具有改进的结合特性的新型mAb变体的方法。使用各种生物信息学工具,我们已经确定了Adalimumab mAb上的重要氨基酸残基。接下来,我们搜索其他残基是否适合突变显着残基,并从合适的突变组合中设计了变体。为了找到最重要的突变,使用分子对接检查和分子动力学模拟将变体的结合特性与野生型阿达木单抗mAb进行了比较。最后,分析了变体和野生型之间的结构特性。我们已经确定了阿达木单抗单克隆抗体上涉及抗原-抗体相互作用的六个最重要的残基。使用合适的突变替换这些残基中的每一个,我们已经建模了143个变体。从数个对接分析中,我们发现了五个重要的变异体,并且在进行分子动力学模拟后,发现了一种具有改善的结合亲和力的最重要的变异体,其结构性质与野生型Adalimumab mAb相似。这项研究设计的变体可能会为单克隆抗体基于结构的亲和力改善提供新的见解,同样,Fc区的修饰也将改善抗体的治疗效应子功能。



























 京公网安备 11010802027423号
京公网安备 11010802027423号