当前位置:
X-MOL 学术
›
Eur. J. Neurosci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Non‐invasive epigenomic molecular phenotyping of the human brain via liquid biopsy of cerebrospinal fluid and next generation sequencing
European Journal of Neuroscience ( IF 2.7 ) Pub Date : 2020-10-06 , DOI: 10.1111/ejn.14997 Lisa Pan 1, 2 , Lora McClain 2 , Patricia Shaw 3 , Nicole Donnellan 3, 4 , Tianjiao Chu 3, 4 , David Finegold 1 , David Peters 1, 2, 3, 4
European Journal of Neuroscience ( IF 2.7 ) Pub Date : 2020-10-06 , DOI: 10.1111/ejn.14997 Lisa Pan 1, 2 , Lora McClain 2 , Patricia Shaw 3 , Nicole Donnellan 3, 4 , Tianjiao Chu 3, 4 , David Finegold 1 , David Peters 1, 2, 3, 4
Affiliation
|
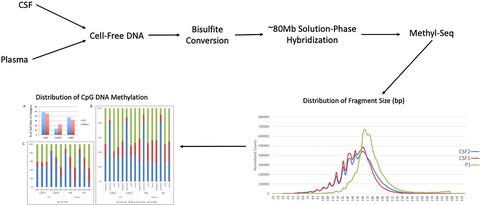
Our goal was to undertake a genome‐wide epigenomic liquid biopsy of cerebrospinal fluid (CSF) for the comprehensive analysis of cell‐free DNA (cfDNA) methylation signatures in the human central nervous system (CNS). Solution‐phase hybridization and massively parallel sequencing of bisulfite converted human DNA was employed to compare methylation signatures of cfDNA obtained from CSF with plasma. Recovery of cfDNA from CSF was relatively low (68–840 pg/mL) compared to plasma (2720–8390 pg/mL) and cfDNA fragments from CSF were approximately 20 bp shorter than their plasma‐derived counterparts. Distributions of CpG methylation signatures were significantly altered between CSF and plasma, both globally and at the level of functional elements including exons, introns, CpG islands, and shores. Sliding window analysis was used to identify differentially methylated regions. We found numerous gene/locus‐specific differences in CpG methylation between cfDNA from CSF and plasma. These loci were more frequently hypomethylated in CSF compared to plasma. Differentially methylated CpGs in CSF were identified in genes related to branching of neurites and neuronal development. Using the GTEx RNA expression database, we found clear association between tissue‐specific gene expression in the CNS and cfDNA methylation patterns in CSF. Ingenuity pathway analysis of differentially methylated regions identified an enrichment of functional pathways related to neurobiology. In conclusion, we present a genome‐wide analysis of DNA methylation in human CSF. Our methods and the resulting data demonstrate the potential of epigenomic liquid biopsy of the human CNS for molecular phenotyping of brain‐derived DNA methylation signatures.
中文翻译:

脑脊液液体活检和下一代测序技术对人脑的非侵入性表观基因组分子表型
我们的目标是对脑脊液(CSF)进行全基因组的表观基因组液体活检,以全面分析人中枢神经系统(CNS)中无细胞DNA(cfDNA)甲基化标记。亚硫酸氢盐转化的人类DNA的溶液相杂交和大规模平行测序被用于比较从CSF获得的cfDNA与血浆的甲基化特征。与血浆(2720–8390 pg / mL)相比,从脑脊液中回收cfDNA(68–840 pg / mL)相对较低,并且从脑脊液中回收cfDNA片段比血浆来源的cfDNA片段短约20 bp。在全球以及在功能元件(包括外显子,内含子,CpG岛和海岸)的水平上,CSF和血浆之间CpG甲基化标记的分布都发生了显着变化。滑动窗口分析用于鉴定差异甲基化区域。我们发现来自脑脊液和血浆的cfDNA之间在CpG甲基化方面存在许多基因/基因座特异性差异。与血浆相比,这些基因座在脑脊液中更经常发生低甲基化。在与神经突分支和神经元发育有关的基因中发现了脑脊液中甲基化的CpGs差异。使用GTEx RNA表达数据库,我们发现CNS中的组织特异性基因表达与CSF中的cfDNA甲基化模式之间存在明显的关联。差异甲基化区域的独创性途径分析确定了与神经生物学相关的功能性途径的丰富化。总之,我们提出了人类CSF中DNA甲基化的全基因组分析。
更新日期:2020-10-26
中文翻译:

脑脊液液体活检和下一代测序技术对人脑的非侵入性表观基因组分子表型
我们的目标是对脑脊液(CSF)进行全基因组的表观基因组液体活检,以全面分析人中枢神经系统(CNS)中无细胞DNA(cfDNA)甲基化标记。亚硫酸氢盐转化的人类DNA的溶液相杂交和大规模平行测序被用于比较从CSF获得的cfDNA与血浆的甲基化特征。与血浆(2720–8390 pg / mL)相比,从脑脊液中回收cfDNA(68–840 pg / mL)相对较低,并且从脑脊液中回收cfDNA片段比血浆来源的cfDNA片段短约20 bp。在全球以及在功能元件(包括外显子,内含子,CpG岛和海岸)的水平上,CSF和血浆之间CpG甲基化标记的分布都发生了显着变化。滑动窗口分析用于鉴定差异甲基化区域。我们发现来自脑脊液和血浆的cfDNA之间在CpG甲基化方面存在许多基因/基因座特异性差异。与血浆相比,这些基因座在脑脊液中更经常发生低甲基化。在与神经突分支和神经元发育有关的基因中发现了脑脊液中甲基化的CpGs差异。使用GTEx RNA表达数据库,我们发现CNS中的组织特异性基因表达与CSF中的cfDNA甲基化模式之间存在明显的关联。差异甲基化区域的独创性途径分析确定了与神经生物学相关的功能性途径的丰富化。总之,我们提出了人类CSF中DNA甲基化的全基因组分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号