当前位置:
X-MOL 学术
›
Phys. Chem. Chem. Phys.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Can modified DNA base pairs with chalcogen bonding expand the genetic alphabet? A combined quantum chemical and molecular dynamics simulation study
Physical Chemistry Chemical Physics ( IF 2.9 ) Pub Date : 2020-10-05 , DOI: 10.1039/d0cp04921b Karan Deep Sharma 1, 2, 3, 4, 5 , Preetleen Kathuria 1, 2, 3, 4, 5 , Stacey D. Wetmore 6, 7, 8 , Purshotam Sharma 1, 2, 3, 4, 5
Physical Chemistry Chemical Physics ( IF 2.9 ) Pub Date : 2020-10-05 , DOI: 10.1039/d0cp04921b Karan Deep Sharma 1, 2, 3, 4, 5 , Preetleen Kathuria 1, 2, 3, 4, 5 , Stacey D. Wetmore 6, 7, 8 , Purshotam Sharma 1, 2, 3, 4, 5
Affiliation
|
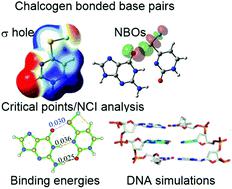
A comprehensive (DFT and MD) computational study is presented with the goal to design and analyze model chalcogen-bonded modified nucleobase pairs that replace one (i.e., AXY:T, G:CXY, GXY:C) or two (GXY:CX′Y′, X/X′ = S, Se and Y/Y′ = F, Cl, Br) Watson–Crick (WC) hydrogen bonds of the canonical A:T or G:C pair with chalcogen bond(s). DFT calculations on 18 base pair combinations that replace one WC hydrogen bond with a chalcogen bond reveal that the bases favorably interact in the gas phase (binding strengths up to −140 kJ mol−1) and water (up to −85 kJ mol−1). Although the remaining hydrogen bond(s) exhibits similar characteristics to those in the canonical base pairs, the structural features of the (Y–X⋯O) chalcogen bond(s) change significantly with the identity of X and Y. The 36 doubly-substituted (GXY:CX′Y′) base pairs have structural deviations from canonical G:C similar to those of the singly-substituted modifications (G:CXY or GXY:C). Furthermore, despite the replacement of two strong hydrogen bonds with chalcogen bonds, some GXY:CX′Y′ pairs possess comparable binding energies (up to −132 kJ mol−1 in the gas phase and up to −92 kJ mol−1 in water) to the most stable G:CXY or GXY:C pairs, as well as canonical G:C. More importantly, G:C-modified pairs containing X = Se (high polarizability) and Y = F (high electronegativity) are the most stable, with comparable or slightly larger (by up to 13 kJ mol−1) binding energies than G:C. Further characterization of the chalcogen bonding in all modified base pairs (AIM, NBO and NCI analyses) reveals that the differences in the binding energies of modified base pairs are mainly dictated by the differences in the strengths of their chalcogen bonds. Finally, MD simulations on DNA oligonucleotides containing the most stable chalcogen-bonded base pair from each of the four classifications (AXY:T, G:CXY, GXY:C and GXY:CX′Y′) reveal that the singly-modified G:C pairs best retain the local helical structure and pairing stability to a greater extent than the modified A:T pair. Overall, our study identifies two (G:CSeF and GSeF:C) promising pairs that retain chalcogen bonding in DNA and should be synthesized and further explored in terms of their potential to expand the genetic alphabet.
中文翻译:

具有硫族元素键的修饰DNA碱基对能否扩展遗传字母?量子化学和分子动力学的组合模拟研究
提出了一项全面的(DFT和MD)计算研究,其目的是设计和分析模型硫族元素键合修饰的碱基对,以取代一个(即A XY:T,G:C XY,G XY:C)或两个(G XY:C X'Y',X / X'= S,Se和Y / Y'= F,Cl,Br)具有硫族元素键的规范A:T或G:C对的沃森克里克(WC)氢键(s)。通过对18个碱基对组合进行DFT计算,用一个硫族元素键取代了一个WC氢键,结果表明这些碱基在气相(结合强度最高为-140 kJ mol -1)和水(最高为-85 kJ mol -1)之间相互作用良好)。尽管剩余的氢键显示出与规范碱基对相似的特征,但(Y–X⋯O)硫族元素键的结构特征随X和Y的身份而发生显着变化。取代的(G XY:C X'Y')碱基对与标准G:C的结构偏差与单取代修饰(G:C XY或G XY:C)相似。此外,尽管用硫族元素键代替了两个强氢键,但某些G XY:C X'Y'对具有相当的结合能(气相中高达-132 kJ mol -1和高达-92 kJ mol -1在水中)至最稳定的G:CXY或G XY:C对,以及规范的G:C。更重要的是,含有X = Se(高极化率)和Y = F(高电负性)的G:C修饰对最稳定,其结合能与G相当或略大(最高13 kJ mol -1): C。在所有修饰碱基对中硫属元素键的进一步表征(AIM,NBO和NCI分析)表明,修饰碱基对的结合能差异主要由其硫属元素键强度差异决定。最后,对来自四个分类(A XY:T,G:C XY,G XY:C和G)中最稳定的硫族元素键合碱基对的DNA寡核苷酸进行MD模拟XY:C X'Y')表明,单修饰的G:C对与修饰的A:T对相比,在更大程度上保留了局部螺旋结构和配对稳定性。总的来说,我们的研究确定了两个(G:C SeF和G SeF:C)有前途的对,它们在DNA中保留了硫族元素键,应该加以合成,并就其扩展遗传字母的潜力进行进一步的探索。
更新日期:2020-10-17
中文翻译:

具有硫族元素键的修饰DNA碱基对能否扩展遗传字母?量子化学和分子动力学的组合模拟研究
提出了一项全面的(DFT和MD)计算研究,其目的是设计和分析模型硫族元素键合修饰的碱基对,以取代一个(即A XY:T,G:C XY,G XY:C)或两个(G XY:C X'Y',X / X'= S,Se和Y / Y'= F,Cl,Br)具有硫族元素键的规范A:T或G:C对的沃森克里克(WC)氢键(s)。通过对18个碱基对组合进行DFT计算,用一个硫族元素键取代了一个WC氢键,结果表明这些碱基在气相(结合强度最高为-140 kJ mol -1)和水(最高为-85 kJ mol -1)之间相互作用良好)。尽管剩余的氢键显示出与规范碱基对相似的特征,但(Y–X⋯O)硫族元素键的结构特征随X和Y的身份而发生显着变化。取代的(G XY:C X'Y')碱基对与标准G:C的结构偏差与单取代修饰(G:C XY或G XY:C)相似。此外,尽管用硫族元素键代替了两个强氢键,但某些G XY:C X'Y'对具有相当的结合能(气相中高达-132 kJ mol -1和高达-92 kJ mol -1在水中)至最稳定的G:CXY或G XY:C对,以及规范的G:C。更重要的是,含有X = Se(高极化率)和Y = F(高电负性)的G:C修饰对最稳定,其结合能与G相当或略大(最高13 kJ mol -1): C。在所有修饰碱基对中硫属元素键的进一步表征(AIM,NBO和NCI分析)表明,修饰碱基对的结合能差异主要由其硫属元素键强度差异决定。最后,对来自四个分类(A XY:T,G:C XY,G XY:C和G)中最稳定的硫族元素键合碱基对的DNA寡核苷酸进行MD模拟XY:C X'Y')表明,单修饰的G:C对与修饰的A:T对相比,在更大程度上保留了局部螺旋结构和配对稳定性。总的来说,我们的研究确定了两个(G:C SeF和G SeF:C)有前途的对,它们在DNA中保留了硫族元素键,应该加以合成,并就其扩展遗传字母的潜力进行进一步的探索。











































 京公网安备 11010802027423号
京公网安备 11010802027423号