Structure ( IF 4.4 ) Pub Date : 2020-10-01 , DOI: 10.1016/j.str.2020.09.003 Michael W Martynowycz 1 , Tamir Gonen 1
|
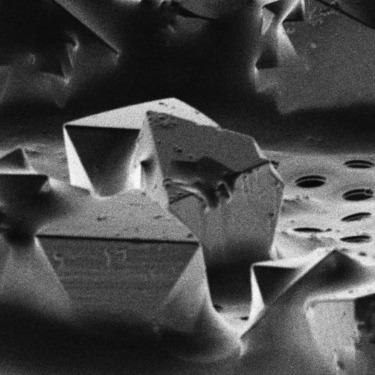
A high throughout method for soaking ligands into protein microcrystals on TEM grids is presented. Every crystal on the grid is soaked simultaneously using only standard cryoelectron microscopy vitrification equipment. The method is demonstrated using proteinase K microcrystals soaked with the 5-amino-2,4,6-triodoisophthalic acid (I3C) magic triangle. A soaked microcrystal is milled to a thickness of approximately 200 nm using a focused ion beam, and MicroED data are collected. A high-resolution structure of the protein with four ligands at high occupancy is determined. Both the number of ligands bound and their occupancy is higher using on-grid soaking of microcrystals compared with much larger crystals treated similarly and investigated by X-ray crystallography. These results indicate that on-grid soaking ligands into microcrystals results in efficient uptake of ligands into protein microcrystals.
中文翻译:

通过在线浸泡将配体掺入用于 MicroED 的蛋白质微晶中
提出了一种将配体浸泡在 TEM 网格上的蛋白质微晶中的高通量方法。仅使用标准低温电子显微镜玻璃化设备同时浸泡网格上的每个晶体。该方法使用用 5-氨基-2,4,6-三碘间苯二甲酸 (I3C) 魔三角浸泡的蛋白酶 K 微晶进行演示。使用聚焦离子束将浸泡过的微晶研磨至约 200 nm 的厚度,并收集 MicroED 数据。确定了在高占有率下具有四个配体的蛋白质的高分辨率结构。与类似处理并通过 X 射线晶体学研究的大得多的晶体相比,使用微晶的网格浸泡,结合的配体数量和它们的占有率都更高。











































 京公网安备 11010802027423号
京公网安备 11010802027423号