Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-10-01 , DOI: 10.1016/j.csbj.2020.09.032 Panupong Mahalapbutr , Napat Kongtaworn , Thanyada Rungrotmongkol
|
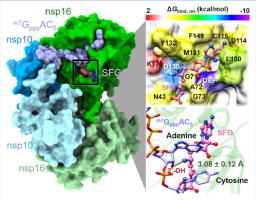
The recent ongoing coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to rapidly spread across the world. To date, neither a specific antiviral drug nor a clinically effective vaccine is available. Among the 15 viral non-structural proteins (nsps), nsp16 methyltransferase has been considered as a potential target due to its crucial role in RNA cap 2’-O-methylation process, preventing the virus detection by cell innate immunity mechanisms. In the present study, molecular recognition between the two natural nucleoside analogs (S-adenosyl-L-homocysteine (SAH) and sinefungin (SFG)) and the SARS-CoV-2 nsp16/nsp10/m7GpppAC5 was studied using all-atom molecular dynamics simulations and free energy calculations based on MM/GBSA and WaterSwap approaches. The binding affinity and the number of hot-spot residues, atomic contacts, and H-bond formations of the SFG/nsp16 complex were distinctly higher than those of the SAH/nsp16 system, consistent with the lower water accessibility at the enzyme active site. Notably, only SFG could electrostatically interact with the 2’-OH and N3 groups of RNA’s adenosine moiety, mimicking the methyl transfer reaction of S-adenosyl-L-methionine substrate. The atomistic binding mechanism obtained from this work paves the way for further optimizations and designs of more specific SARS-CoV-2 nsp16 inhibitors in the fight against COVID-19.
中文翻译:

识别SARS-CoV-2 Nsp16 / Nsp10 RNA Cap 2'-O-甲基转移酶中S-腺苷-L-高半胱氨酸和新氟菌素结构的洞察力
由严重的急性呼吸系统综合症冠状病毒2(SARS-CoV-2)引起的最近正在进行的2019年冠状病毒病(COVID-19)大流行继续在世界范围内迅速蔓延。迄今为止,既没有特定的抗病毒药物,也没有临床有效的疫苗。在15种病毒非结构蛋白(nsps)中,由于nsp16甲基转移酶在RNA cap 2'-O-甲基化过程中起着至关重要的作用,因此被认为是潜在的靶标,它通过细胞固有的免疫机制阻止了病毒的检测。在本研究中,两种天然核苷类似物(S-腺苷-L-同型半胱氨酸(SAH)和西芬芬净(SFG))与SARS-CoV-2 nsp16 / nsp10 / m7 G ppp AC 5之间的分子识别使用基于MM / GBSA和WaterSwap方法的全原子分子动力学模拟和自由能计算研究了碳纳米管。SFG / nsp16复合物的结合亲和力和热点残基,原子接触和H键形成的数量明显高于SAH / nsp16系统的结合亲和力,这与酶活性位点的水可及性较低有关。值得注意的是,只有SFG可以与RNA腺苷部分的2'-OH和N3基团发生静电相互作用,从而模仿S-腺苷-L-蛋氨酸的甲基转移反应。从这项工作中获得的原子结合机制为进一步优化和设计更特异性的SARS-CoV-2 nsp16抑制剂与COVID-19的斗争铺平了道路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号