Cell Chemical Biology ( IF 6.6 ) Pub Date : 2020-10-01 , DOI: 10.1016/j.chembiol.2020.09.005 Jonathan W Bushman 1 , Katherine A Donovan 1 , Nathan J Schauer 1 , Xiaoxi Liu 1 , Wanyi Hu 1 , Anthony C Varca 1 , Sara J Buhrlage 1 , Eric S Fischer 1
|
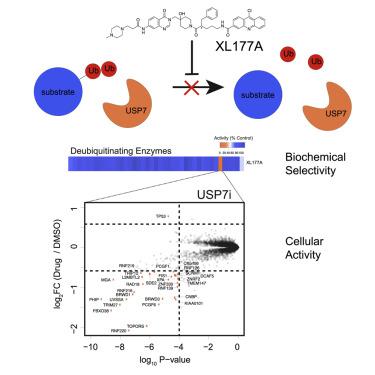
Deubiquitinating enzymes (DUBs) catalyze the removal of ubiquitin, thereby reversing the activity of E3 ubiquitin ligases and are central to the control of protein abundance and function. Despite the growing interest in DUBs as therapeutic targets, cellular functions for DUBs remain largely unknown and technical challenges often preclude the identification of DUB substrates in a comprehensive manner. Here, we demonstrate that treatment with potent DUB inhibitors coupled to mass spectrometry-based proteomics can identify DUB substrates at a proteome-wide scale. We applied this approach to USP7, a DUB widely investigated as a therapeutic target and identified many known substrates and additional targets. We demonstrate that USP7 substrates are enriched for DNA repair enzymes and E3 ubiquitin ligases. This work provides not only a comprehensive annotation of USP7 substrates, but a general protocol widely applicable to other DUBs, which is critical for translational development of DUB targeted agents.
中文翻译:

使用选择性抑制剂基于蛋白质组学鉴定 DUB 底物
去泛素化酶 (DUB) 催化泛素的去除,从而逆转 E3 泛素连接酶的活性,是控制蛋白质丰度和功能的核心。尽管人们对 DUB 作为治疗靶点的兴趣日益浓厚,但 DUB 的细胞功能在很大程度上仍是未知的,技术挑战通常会妨碍对 DUB 底物的全面鉴定。在这里,我们证明了有效的 DUB 抑制剂与基于质谱的蛋白质组学相结合的治疗可以在蛋白质组范围内识别 DUB 底物。我们将这种方法应用于 USP7,这是一种作为治疗靶点被广泛研究的 DUB,并确定了许多已知的底物和其他靶点。我们证明 USP7 底物富含 DNA 修复酶和 E3 泛素连接酶。











































 京公网安备 11010802027423号
京公网安备 11010802027423号