Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-11-30 , DOI: 10.2174/1574893615666200224095531 Zhihua Du 1 , Xiangdong Xiao 1 , Vladimir N. Uversky 2
|
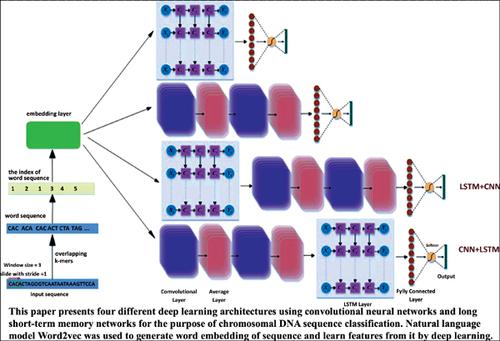
Background: Chromosomal DNA contains most of the genetic information of eukaryotes and plays an important role in the growth, development and reproduction of living organisms. Most chromosomal DNA sequences are known to wrap around histones, and distinguishing these DNA sequences from ordinary DNA sequences is important for understanding the genetic code of life. The main difficulty behind this problem is the feature selection process. DNA sequences have no explicit features, and the common representation methods, such as onehot coding, introduced the major drawback of high dimensionality. Recently, deep learning models have been proved to be able to automatically extract useful features from input patterns.
Objective: We aim to investigate which deep learning networks could achieve notable improvements in the field of DNA sequence classification using only sequence information. Methods: In this paper, we present four different deep learning architectures using convolutional neural networks and long short-term memory networks for the purpose of chromosomal DNA sequence classification. Natural language model Word2vec was used to generate word embedding of sequence and learn features from it by deep learning.
Results: The comparison of these four architectures is carried out on 10 chromosomal DNA datasets. The results show that the architecture of convolutional neural networks combined with long short-term memory networks is superior to other methods with regards to the accuracy of chromosomal DNA prediction.
Conclusion: In this study, four deep learning models were compared for an automatic classification of chromosomal DNA sequences with no steps of sequence preprocessing. In particular, we have regarded DNA sequences as natural language and extracted word embedding with Word2Vec to represent DNA sequences. Results show a superiority of the CNN+LSTM model in the ten classification tasks. The reason for this success is that the CNN module captures the regulatory motifs, while the following LSTM layer captures the long-term dependencies between them.
中文翻译:

使用混合深度学习架构对染色体DNA序列进行分类
背景:染色体DNA包含真核生物的大多数遗传信息,并且在生物体的生长,发育和繁殖中发挥重要作用。已知大多数染色体DNA序列会包裹组蛋白,将这些DNA序列与普通DNA序列区分开对于理解生命的遗传密码很重要。这个问题背后的主要困难是特征选择过程。DNA序列没有明确的特征,常用的表示方法(如onehot编码)引入了高维数的主要缺点。最近,事实证明,深度学习模型能够从输入模式中自动提取有用的功能。
目的:我们旨在研究仅在序列信息的基础上,哪些深度学习网络可以在DNA序列分类领域取得显着改善。方法:在本文中,我们提出了四种使用卷积神经网络和长短期记忆网络的深度学习架构,用于染色体DNA序列分类。自然语言模型Word2vec用于生成序列的单词嵌入,并通过深度学习从序列中学习特征。
结果:这四种结构的比较是在10个染色体DNA数据集上进行的。结果表明,卷积神经网络与长短期记忆网络相结合的架构在染色体DNA预测的准确性方面优于其他方法。
结论:在这项研究中,比较了四种深度学习模型,它们无需进行序列预处理即可自动分类染色体DNA序列。特别是,我们将DNA序列视为自然语言,并提取了用Word2Vec嵌入的单词来表示DNA序列。结果显示了CNN + LSTM模型在十个分类任务中的优越性。成功的原因是CNN模块捕获了调控基元,而随后的LSTM层捕获了它们之间的长期依赖性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号