Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-11-30 , DOI: 10.2174/1574893615666200224095041 Jianwei Li 1 , Leibo Liu 1 , Qinghua Cui 2 , Yuan Zhou 2
|
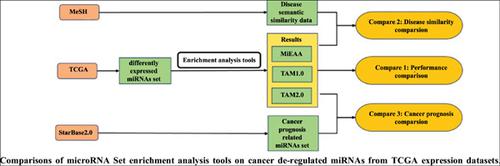
Background: De-regulation of microRNAs (miRNAs) is closely related to many complex diseases, including cancers. In The Cancer Genome Atlas (TCGA), hundreds of differentially expressed miRNAs are stored for each type of cancer, which are hard to be intuitively interpreted. To date, several miRNA set enrichment tools have been tailored to predict the potential disease associations and functions of de-regulated miRNAs, including the miRNA Enrichment Analysis and Annotation tool (miEAA) and Tool for Annotations of human MiRNAs (TAM1.0 &TAM 2.0). However, independent benchmarking of these tools is warranted to assess their effectiveness and robustness, and the relationship between enrichment analysis results and the prognosis significance of cancers.
Methods: Based on differentially expressed miRNAs from expression profiles in TCGA, we performed a series of tests and a comprehensive comparison of the enrichment analysis results of miEAA, TAM 1.0 and TAM 2.0. The work focused on the performance of the three tools, disease similarity based on miRNA-disease associations from the enrichment analysis results, the relationship between the overrepresented miRNAs from enrichment analysis results and the prognosis significance of cancers.
Results: The main results show that TAM 2.0 is more likely to identify the regulatory disease’s functions of de-regulated miRNA; it is feasible to calculate disease similarity based on enrichment analysis results of TAM 2.0; and there is weak positive correlation between the occurrence frequency of miRNAs in the TAM 2.0 enrichment analysis results and the prognosis significance of the cancer miRNAs.
Conclusion: Our comparison results not only provide a reference for biomedical researchers to choose appropriate miRNA set enrichment analysis tools to achieve their purpose but also demonstrate that the degree of overrepresentation of miRNAs could be a supplementary indicator of the disease similarity and the prognostic effect of cancer miRNAs.
中文翻译:

来自TCGA表达数据集的针对癌症失控的miRNA的MicroRNA集富集分析工具的比较
背景:microRNA(miRNA)的失调与许多复杂的疾病(包括癌症)密切相关。在《癌症基因组图谱》(TCGA)中,每种癌症都存储了数百种差异表达的miRNA,很难直观地解释。迄今为止,已经对几种miRNA集富集工具进行了量身定制,以预测失调的miRNA的潜在疾病关联和功能,包括miRNA富集分析和注释工具(miEAA)和人类MiRNA注释工具(TAM1.0&TAM 2.0) 。但是,必须对这些工具进行独立的基准测试,以评估其有效性和鲁棒性,以及富集分析结果与癌症预后意义之间的关系。
方法:基于TCGA表达谱中差异表达的miRNA,我们进行了一系列测试并对miEAA,TAM 1.0和TAM 2.0的富集分析结果进行了全面比较。这项工作着眼于这三种工具的性能,基于富集分析结果中基于miRNA-疾病关联的疾病相似性,基于富集分析结果中所代表的miRNA之间的关系以及癌症的预后意义。
结果:主要结果表明,TAM 2.0更有可能鉴定出调节性miRNA失调的调节性疾病功能。根据TAM 2.0的富集分析结果计算疾病相似度是可行的。TAM 2.0富集分析结果中miRNA的出现频率与癌症miRNA的预后意义之间呈弱正相关。
结论:我们的比较结果不仅为生物医学研究人员选择合适的miRNA集富集分析工具以实现其目的提供了参考,而且还证明了miRNA过度表达的程度可以作为疾病相似性和癌症预后效果的补充指标miRNA。











































 京公网安备 11010802027423号
京公网安备 11010802027423号