Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-10-31 , DOI: 10.2174/1574893615999200414093636 Pan Wang 1 , Guiyang Zhang 1 , You Li 1 , Ammar Oad 1 , Guohua Huang 1
|
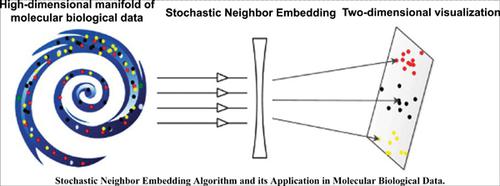
With the advent of the era of big data, the numbers and the dimensions of data are increasingly becoming larger. It is very critical to reduce dimensions or visualize data and then uncover the hidden patterns of characteristics or the mechanism underlying data. Stochastic Neighbor Embedding (SNE) has been developed for data visualization over the last ten years. Due to its efficiency in the visualization of data, SNE has been applied to a wide range of fields. We briefly reviewed the SNE algorithm and its variants, summarizing application of it in visualizing single-cell sequencing data, single nucleotide polymorphisms, and mass spectrometry imaging data. We also discussed the strength and the weakness of the SNE, with a special emphasis on how to set parameters to promote quality of visualization, and finally indicated potential development of SNE in the coming future.
中文翻译:

随机邻域嵌入算法及其在分子生物学数据中的应用
随着大数据时代的到来,数据的数量和维度越来越大。缩小尺寸或可视化数据,然后发现特征的隐藏模式或数据背后的机制非常关键。在过去的十年中,随机邻居嵌入(SNE)已开发用于数据可视化。由于其在数据可视化方面的效率,SNE已被广泛应用于各个领域。我们简要回顾了SNE算法及其变体,总结了其在可视化单细胞测序数据,单核苷酸多态性和质谱成像数据中的应用。我们还讨论了SNE的优势和劣势,特别着重于如何设置参数以提高可视化质量,











































 京公网安备 11010802027423号
京公网安备 11010802027423号