Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-11-30 , DOI: 10.2174/1574893615666200316101205 Hongdong Li 1 , Wenjing Zhang 2 , Yuwen Luo 3 , Jianxin Wang 1
|
Background: Transcriptome annotation is the basis for understanding gene structures and analysing gene expression. The transcriptome annotation of many organisms such as humans is far from incomplete, due partly to the challenge in the identification of isoforms that are produced from the same gene through alternative splicing. Third generation sequencing (TGS) reads provide unprecedented opportunity for detecting isoforms due to their long length that exceeds the length of most isoforms. One limitation of current TGS reads-based isoform detection methods is that they are exclusively based on sequence reads, without incorporating the sequence information of annotated isoforms.
Objective: We aim to develop a method to detect isoforms by incorporating annotated isoforms.
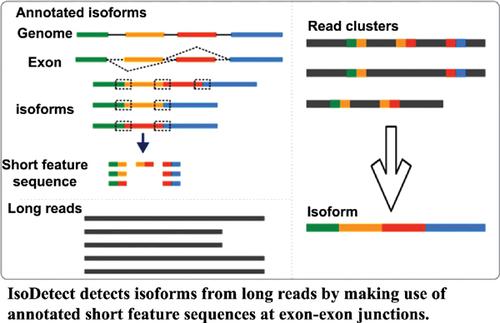
Methods: Based on annotated isoforms, we propose a splice isoform detection method called IsoDetect. First, the sequence at exon-exon junctions is extracted from annotated isoforms as “short feature sequences”, which is used to distinguish splice isoforms. Second, we align these feature sequences to long reads and partition long reads into groups that contain the same set of feature sequences, thereby avoiding the pair-wise comparison among the large number of long reads. Third, clustering and consensus generation are carried out based on sequence similarity. For the long reads that do not contain any short feature sequence, clustering analysis based on sequence similarity is performed to identify isoforms. Therefore, our method can detect not only known but also novel isoforms.
Results: Tested on two datasets from Calypte anna and Zebra Finch, IsoDetect shows higher speed and good accuracies compared with four existing methods.
Conclusion: IsoDetect may become a promising method for isoform detection.
中文翻译:

IsoDetect:基于短特征序列从第三代长读中检测剪接同工型
背景:转录组注释是理解基因结构和分析基因表达的基础。许多生物(例如人类)的转录组注释远非完整,部分原因是在鉴定通过替代剪接从同一基因产生的同工型方面存在挑战。第三代测序(TGS)读段提供了前所未有的机会来检测同工型,因为它们的长度超过了大多数同工型的长度。当前基于TGS读数的同工型检测方法的一个局限性在于,它们仅基于序列读数,而没有并入带注释的同工型的序列信息。
目的:我们旨在开发一种通过结合注释的同工型检测同工型的方法。
方法:基于带注释的同工型,我们提出了一种称为IsoDetect的剪接同工型检测方法。首先,从注释的同工型中提取外显子-外显子连接处的序列作为“短特征序列”,用于区分剪接同工型。其次,我们将这些特征序列与长读段对齐,并将长读段划分为包含相同特征序列集的组,从而避免了大量长读段之间的成对比较。第三,基于序列相似性进行聚类和共识生成。对于不包含任何短特征序列的长读段,基于序列相似性进行聚类分析以鉴定同工型。因此,我们的方法不仅可以检测已知的,而且还可以检测新的同工型。
结果:IsoDetect在来自Calypte anna和Zebra Finch的两个数据集上进行了测试,与四种现有方法相比,具有更高的速度和更高的准确性。
结论:IsoDetect可能成为有前途的异构体检测方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号