Current Bioinformatics ( IF 4 ) Pub Date : 2020-10-31 , DOI: 10.2174/1574893615666200120103948 Xiaojuan Yu 1 , Jianguo Zhou 1 , Mingming Zhao 1 , Chao Yi 1 , Qing Duan 1 , Wei Zhou 1 , Jin Li 1
|
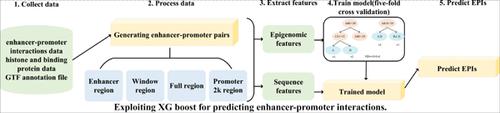
Background: Gene expression and disease control are regulated by the interaction between distal enhancers and proximal promoters, and the study of enhancer promoter interactions (EPIs) provides insight into the genetic basis of diseases.
Objective: Although the recent emergence of high-throughput sequencing methods have a deepened understanding of EPIs, accurate prediction of EPIs still limitations.
Methods: We have implemented a XGBoost-based approach and introduced two sets of features (epigenomic and sequence) to predict the interactions between enhancers and promoters in different cell lines.
Results: Extensive experimental results show that XGBoost effectively predicts EPIs across three cell lines, especially when using epigenomic and sequence features.
Conclusion: XGBoost outperforms other methods, such as random forest, Adadboost, GBDT, and TargetFinder.
中文翻译:

利用XG Boost预测增强剂与促进剂的相互作用
背景:基因表达和疾病控制受远端增强子和近端启动子之间的相互作用的调节,增强子启动子相互作用(EPI)的研究为了解疾病的遗传基础提供了见识。
目的:尽管最近出现的高通量测序方法已经加深了对EPI的理解,但准确预测EPI仍然存在局限性。
方法:我们已经实现了基于XGBoost的方法,并引入了两组功能(表基因组和序列)来预测不同细胞系中增强子和启动子之间的相互作用。
结果:广泛的实验结果表明,XGBoost可有效预测三种细胞系的EPI,特别是在使用表观基因组和序列特征时。
结论:XGBoost优于其他方法,例如随机森林,Adadboost,GBDT和TargetFinder。



























 京公网安备 11010802027423号
京公网安备 11010802027423号