Current Bioinformatics ( IF 2.4 ) Pub Date : 2020-06-30 , DOI: 10.2174/1574893614666190902151038 Xiao-Fei Yang 1 , Yuan-Ke Zhou 1 , Lin Zhang 1 , Yang Gao 2 , Pu-Feng Du 1
|
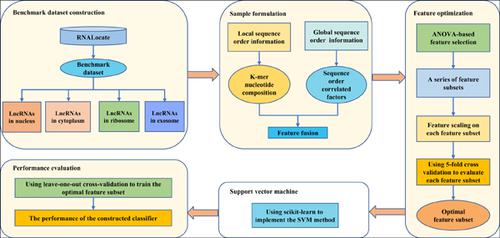
Background: Long non-coding RNAs (lncRNAs) are transcripts with a length more than 200 nucleotides, functioning in the regulation of gene expression. More evidence has shown that the biological functions of lncRNAs are intimately related to their subcellular localizations. Therefore, it is very important to confirm the lncRNA subcellular localization.
Methods: In this paper, we proposed a novel method to predict the subcellular localization of lncRNAs. To more comprehensively utilize lncRNA sequence information, we exploited both kmer nucleotide composition and sequence order correlated factors of lncRNA to formulate lncRNA sequences. Meanwhile, a feature selection technique which was based on the Analysis Of Variance (ANOVA) was applied to obtain the optimal feature subset. Finally, we used the support vector machine (SVM) to perform the prediction.
Results: The AUC value of the proposed method can reach 0.9695, which indicated the proposed predictor is an efficient and reliable tool for determining lncRNA subcellular localization. Furthermore, the predictor can reach the maximum overall accuracy of 90.37% in leave-one-out cross validation, which clearly outperforms the existing state-of- the-art method.
Conclusion: It is demonstrated that the proposed predictor is feasible and powerful for the prediction of lncRNA subcellular. To facilitate subsequent genetic sequence research, we shared the source code at https://github.com/NicoleYXF/lncRNA.
中文翻译:

使用不平衡的伪-k核苷酸组成预测LncRNA亚细胞定位。
背景:长非编码RNA(lncRNA)是长度超过200个核苷酸的转录本,可调控基因表达。更多证据表明,lncRNA的生物学功能与其亚细胞定位密切相关。因此,确认lncRNA亚细胞定位非常重要。
方法:本文提出了一种预测lncRNAs亚细胞定位的新方法。为了更全面地利用lncRNA序列信息,我们利用了lncRNA的kmer核苷酸组成和序列顺序相关因子来制定lncRNA序列。同时,应用了基于方差分析(ANOVA)的特征选择技术来获得最佳特征子集。最后,我们使用支持向量机(SVM)进行预测。
结果:所提出方法的AUC值可达到0.9695,表明所提出的预测因子是确定lncRNA亚细胞定位的有效且可靠的工具。此外,在留一法交叉验证中,预测器可以达到90.37%的最大总体准确度,明显优于现有的最新方法。
结论:表明所提出的预测子对于预测lncRNA亚细胞是可行且强大的。为了方便后续的基因序列研究,我们在https://github.com/NicoleYXF/lncRNA上共享了源代码。











































 京公网安备 11010802027423号
京公网安备 11010802027423号