当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Rosetta-Enabled Structural Prediction of Permissive Loop Insertion Sites in Proteins
Biochemistry ( IF 2.9 ) Pub Date : 2020-09-24 , DOI: 10.1021/acs.biochem.0c00533 Joseph G. Plaks 1 , Jeff A. Brewer 1 , Nicole K. Jacobsen 1 , Michael McKenna 1 , Joshua R. Uzarski 2 , Timothy J. Lawton 2 , Shaun F. Filocamo 2 , Joel L. Kaar 1
Biochemistry ( IF 2.9 ) Pub Date : 2020-09-24 , DOI: 10.1021/acs.biochem.0c00533 Joseph G. Plaks 1 , Jeff A. Brewer 1 , Nicole K. Jacobsen 1 , Michael McKenna 1 , Joshua R. Uzarski 2 , Timothy J. Lawton 2 , Shaun F. Filocamo 2 , Joel L. Kaar 1
Affiliation
|
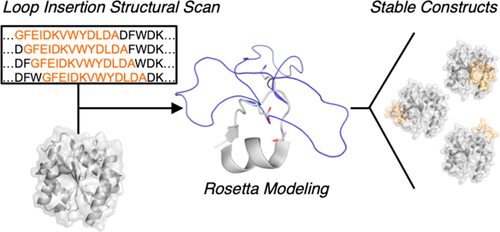
While loop motifs frequently play a major role in protein function, our understanding of how to rationally engineer proteins with novel loop domains remains limited. In the absence of rational approaches, the incorporation of loop domains often destabilizes proteins, thereby requiring massive screening and selection to identify sites that can accommodate loop insertion. We developed a computational strategy for rapidly scanning the entire structure of a scaffold protein to determine the impact of loop insertion at all possible amino acid positions. This approach is based on the Rosetta kinematic loop modeling protocol and was demonstrated by identifying sites in lipase that were permissive to insertion of the LAP peptide. Interestingly, the identification of permissive sites was dependent on the contribution of the residues in the near-loop environment on the Rosetta score and did not correlate with conventional structural features (e.g., B-factors). As evidence of this, several insertion sites (e.g., following residues 17, 47–49, and 108), which were predicted and confirmed to be permissive, interrupted helices, while others (e.g., following residues 43, 67, 116, 119, and 121), which are situated in loop regions, were nonpermissive. This approach was further shown to be predictive for β-glucosidase and human phosphatase and tensin homologue (PTEN), and to facilitate the engineering of insertion sites through in silico mutagenesis. By enabling the design of loop-containing protein libraries with high probabilities of soluble expression, this approach has broad implications in many areas of protein engineering, including antibody design, improving enzyme activity, and protein modification.
中文翻译:

基于Rosetta的蛋白质中容许环插入位点的结构预测
虽然环基序经常在蛋白质功能中起主要作用,但我们对如何合理地改造具有新型环结构域的蛋白质的了解仍然有限。在缺乏合理方法的情况下,环结构域的掺入常常使蛋白质不稳定,从而需要大量筛选和选择以鉴定可容纳环插入的位点。我们开发了一种计算策略,用于快速扫描支架蛋白的整个结构,以确定在所有可能的氨基酸位置处环插入的影响。该方法基于Rosetta运动回路建模协议,并通过识别脂肪酶中允许插入LAP肽的位点得到证明。有趣的是B因子)。对此的证据是,几个插入位点(例如,在残基17、47-49和108之后)被预测并证实是允许的,中断了螺旋,而其他插入位点(例如,在残基43、67、116、119之后,和121)位于环路区域,是不允许的。该方法进一步证明可预测β-葡萄糖苷酶,人磷酸酶和张力蛋白同源物(PTEN),并通过计算机诱变促进插入位点的工程化。通过使设计具有高可溶性表达可能性的含环蛋白质文库成为可能,这种方法在蛋白质工程的许多领域具有广泛的意义,包括抗体设计,提高酶活性和蛋白质修饰。
更新日期:2020-10-21
中文翻译:

基于Rosetta的蛋白质中容许环插入位点的结构预测
虽然环基序经常在蛋白质功能中起主要作用,但我们对如何合理地改造具有新型环结构域的蛋白质的了解仍然有限。在缺乏合理方法的情况下,环结构域的掺入常常使蛋白质不稳定,从而需要大量筛选和选择以鉴定可容纳环插入的位点。我们开发了一种计算策略,用于快速扫描支架蛋白的整个结构,以确定在所有可能的氨基酸位置处环插入的影响。该方法基于Rosetta运动回路建模协议,并通过识别脂肪酶中允许插入LAP肽的位点得到证明。有趣的是B因子)。对此的证据是,几个插入位点(例如,在残基17、47-49和108之后)被预测并证实是允许的,中断了螺旋,而其他插入位点(例如,在残基43、67、116、119之后,和121)位于环路区域,是不允许的。该方法进一步证明可预测β-葡萄糖苷酶,人磷酸酶和张力蛋白同源物(PTEN),并通过计算机诱变促进插入位点的工程化。通过使设计具有高可溶性表达可能性的含环蛋白质文库成为可能,这种方法在蛋白质工程的许多领域具有广泛的意义,包括抗体设计,提高酶活性和蛋白质修饰。











































 京公网安备 11010802027423号
京公网安备 11010802027423号