当前位置:
X-MOL 学术
›
ACS Chem. Neurosci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Pain Chemogenomics Knowledgebase (Pain-CKB) for Systems Pharmacology Target Mapping and PBPK Modeling Investigation of Opioid Drug-Drug Interactions.
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2020-09-23 , DOI: 10.1021/acschemneuro.0c00372 Mingzhe Shen 1 , Maozi Chen 1 , Tianjian Liang 1 , Siyi Wang 1 , Ying Xue 2 , Richard Bertz 1 , Xiang-Qun Xie 3 , Zhiwei Feng 1
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2020-09-23 , DOI: 10.1021/acschemneuro.0c00372 Mingzhe Shen 1 , Maozi Chen 1 , Tianjian Liang 1 , Siyi Wang 1 , Ying Xue 2 , Richard Bertz 1 , Xiang-Qun Xie 3 , Zhiwei Feng 1
Affiliation
|
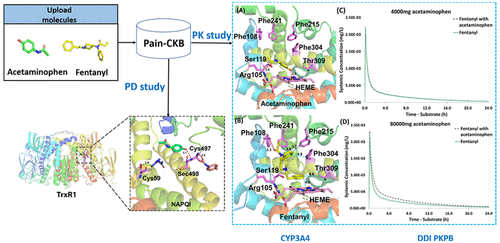
More than 50 million adults in America suffer from chronic pain. Opioids are commonly prescribed for their effectiveness in relieving many types of pain. However, excessive prescribing of opioids can lead to abuse, addiction, and death. Non-steroidal anti-inflammatory drugs (NSAIDs), another major class of analgesic, also have many problematic side effects including headache, dizziness, vomiting, diarrhea, nausea, constipation, reduced appetite, and drowsiness. There is an urgent need for the understanding of molecular mechanisms that underlie drug abuse and addiction to aid in the design of new preventive or therapeutic agents for pain management. To facilitate pain related small-molecule signaling pathway studies and the prediction of potential therapeutic target(s) for the treatment of pain, we have constructed a comprehensive platform of a pain domain-specific chemogenomics knowledgebase (Pain-CKB) with integrated data mining computing tools. Our new computing platform describes the chemical molecules, genes, proteins, and signaling pathways involved in pain regulation. Pain-CKB is implemented with a friendly user interface for the prediction of the relevant protein targets and analysis and visualization of the outputs, including HTDocking, TargetHunter, BBB predictor, and Spider Plot. Combining these with other novel tools, we performed three case studies to systematically demonstrate how further studies can be conducted based on the data generated from Pain-CKB and its algorithms and tools. First, systems pharmacology target mapping was carried out for four FDA approved analgesics in order to identify the known target and predict off-target interactions. Subsequently, the target mapping outcomes were applied to build physiologically based pharmacokinetic (PBPK) models for acetaminophen and fentanyl to explore the drug–drug interaction (DDI) between this pair of drugs. Finally, pharmaco-analytics was conducted to explore the detailed interaction pattern of acetaminophen reactive metabolite and its hepatotoxicity target, thioredoxin reductase.
中文翻译:

用于阿片类药物相互作用的系统药理学靶标定位和 PBPK 建模研究的疼痛化学基因组学知识库 (Pain-CKB)。
美国有超过 5000 万成年人患有慢性疼痛。阿片类药物通常因其能有效缓解多种类型的疼痛而被开出处方。然而,过量服用阿片类药物可能会导致滥用、成瘾和死亡。非甾体类抗炎药 (NSAID) 是另一类主要的镇痛药,也有许多副作用,包括头痛、头晕、呕吐、腹泻、恶心、便秘、食欲下降和嗜睡。迫切需要了解药物滥用和成瘾的分子机制,以帮助设计新的疼痛管理预防或治疗药物。为了促进疼痛相关的小分子信号通路研究和预测治疗疼痛的潜在治疗靶点,我们构建了疼痛领域特定化学基因组学知识库(Pain-CKB)的综合平台,并集成了数据挖掘计算工具。我们的新计算平台描述了参与疼痛调节的化学分子、基因、蛋白质和信号通路。Pain-CKB 通过友好的用户界面实现,用于预测相关蛋白质靶标并分析和可视化输出,包括 HTDocking、TargetHunter、BBB 预测器和 Spider Plot。将这些与其他新颖工具相结合,我们进行了三个案例研究,系统地展示了如何基于 Pain-CKB 及其算法和工具生成的数据进行进一步的研究。第一的,对 FDA 批准的四种镇痛药进行了系统药理学靶点定位,以确定已知靶点并预测脱靶相互作用。随后,目标映射结果被应用于建立对乙酰氨基酚和芬太尼的生理药代动力学(PBPK)模型,以探索这对药物之间的药物相互作用(DDI)。最后,进行药物分析以探索对乙酰氨基酚反应性代谢物与其肝毒性靶标硫氧还蛋白还原酶的详细相互作用模式。
更新日期:2020-10-21
中文翻译:

用于阿片类药物相互作用的系统药理学靶标定位和 PBPK 建模研究的疼痛化学基因组学知识库 (Pain-CKB)。
美国有超过 5000 万成年人患有慢性疼痛。阿片类药物通常因其能有效缓解多种类型的疼痛而被开出处方。然而,过量服用阿片类药物可能会导致滥用、成瘾和死亡。非甾体类抗炎药 (NSAID) 是另一类主要的镇痛药,也有许多副作用,包括头痛、头晕、呕吐、腹泻、恶心、便秘、食欲下降和嗜睡。迫切需要了解药物滥用和成瘾的分子机制,以帮助设计新的疼痛管理预防或治疗药物。为了促进疼痛相关的小分子信号通路研究和预测治疗疼痛的潜在治疗靶点,我们构建了疼痛领域特定化学基因组学知识库(Pain-CKB)的综合平台,并集成了数据挖掘计算工具。我们的新计算平台描述了参与疼痛调节的化学分子、基因、蛋白质和信号通路。Pain-CKB 通过友好的用户界面实现,用于预测相关蛋白质靶标并分析和可视化输出,包括 HTDocking、TargetHunter、BBB 预测器和 Spider Plot。将这些与其他新颖工具相结合,我们进行了三个案例研究,系统地展示了如何基于 Pain-CKB 及其算法和工具生成的数据进行进一步的研究。第一的,对 FDA 批准的四种镇痛药进行了系统药理学靶点定位,以确定已知靶点并预测脱靶相互作用。随后,目标映射结果被应用于建立对乙酰氨基酚和芬太尼的生理药代动力学(PBPK)模型,以探索这对药物之间的药物相互作用(DDI)。最后,进行药物分析以探索对乙酰氨基酚反应性代谢物与其肝毒性靶标硫氧还蛋白还原酶的详细相互作用模式。











































 京公网安备 11010802027423号
京公网安备 11010802027423号