当前位置:
X-MOL 学术
›
Chem. Soc. Rev.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Design of small molecules targeting RNA structure from sequence.
Chemical Society Reviews ( IF 40.4 ) Pub Date : 2020-09-16 , DOI: 10.1039/d0cs00455c Andrei Ursu 1 , Jessica L Childs-Disney 1 , Ryan J Andrews 2 , Collin A O'Leary 2 , Samantha M Meyer 1 , Alicia J Angelbello 1 , Walter N Moss 2 , Matthew D Disney 1
Chemical Society Reviews ( IF 40.4 ) Pub Date : 2020-09-16 , DOI: 10.1039/d0cs00455c Andrei Ursu 1 , Jessica L Childs-Disney 1 , Ryan J Andrews 2 , Collin A O'Leary 2 , Samantha M Meyer 1 , Alicia J Angelbello 1 , Walter N Moss 2 , Matthew D Disney 1
Affiliation
|
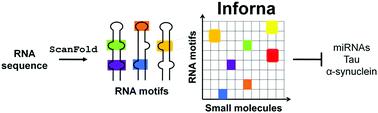
The design and discovery of small molecule medicines has largely been focused on a small number of druggable protein families. A new paradigm is emerging, however, in which small molecules exert a biological effect by interacting with RNA, both to study human disease biology and provide lead therapeutic modalities. Due to this potential for expanding target pipelines and treating a larger number of human diseases, robust platforms for the rational design and optimization of small molecules interacting with RNAs (SMIRNAs) are in high demand. This review highlights three major pillars in this area. First, the transcriptome-wide identification and validation of structured RNA elements, or motifs, within disease-causing RNAs directly from sequence is presented. Second, we provide an overview of high-throughput screening approaches to identify SMIRNAs as well as discuss the lead identification strategy, Inforna, which decodes the three-dimensional (3D) conformation of RNA motifs with small molecule binding partners, directly from sequence. An emphasis is placed on target validation methods to study the causality between modulating the RNA motif in vitro and the phenotypic outcome in cells. Third, emergent modalities that convert occupancy-driven mode of action SMIRNAs into event-driven small molecule chemical probes, such as RNA cleavers and degraders, are presented. Finally, the future of the small molecule RNA therapeutics field is discussed, as well as hurdles to overcome to develop potent and selective RNA-centric chemical probes.
中文翻译:

根据序列设计针对 RNA 结构的小分子。
小分子药物的设计和发现主要集中在少数可成药的蛋白质家族。然而,一种新的范式正在出现,其中小分子通过与 RNA 相互作用发挥生物效应,既可以研究人类疾病生物学,也可以提供领先的治疗方式。由于具有扩大靶标管道和治疗更多人类疾病的潜力,因此迫切需要用于合理设计和优化与 RNA 相互作用的小分子 (SMIRNA) 的强大平台。本次审查强调了该领域的三大支柱。首先,直接从序列中对致病 RNA 中的结构化 RNA 元件或基序进行全转录组鉴定和验证。其次,我们概述了识别 SMIRNA 的高通量筛选方法,并讨论了先导识别策略 Infona,该策略直接从序列中解码具有小分子结合伴侣的 RNA 基序的三维 (3D) 构象。重点放在靶标验证方法上,以研究体外调节 RNA 基序与细胞表型结果之间的因果关系。第三,提出了将占用驱动的作用模式 SMIRNA 转化为事件驱动的小分子化学探针(例如 RNA 切割器和降解器)的新兴模式。最后,讨论了小分子 RNA 治疗领域的未来,以及开发有效且选择性的以 RNA 为中心的化学探针需要克服的障碍。
更新日期:2020-10-19
中文翻译:

根据序列设计针对 RNA 结构的小分子。
小分子药物的设计和发现主要集中在少数可成药的蛋白质家族。然而,一种新的范式正在出现,其中小分子通过与 RNA 相互作用发挥生物效应,既可以研究人类疾病生物学,也可以提供领先的治疗方式。由于具有扩大靶标管道和治疗更多人类疾病的潜力,因此迫切需要用于合理设计和优化与 RNA 相互作用的小分子 (SMIRNA) 的强大平台。本次审查强调了该领域的三大支柱。首先,直接从序列中对致病 RNA 中的结构化 RNA 元件或基序进行全转录组鉴定和验证。其次,我们概述了识别 SMIRNA 的高通量筛选方法,并讨论了先导识别策略 Infona,该策略直接从序列中解码具有小分子结合伴侣的 RNA 基序的三维 (3D) 构象。重点放在靶标验证方法上,以研究体外调节 RNA 基序与细胞表型结果之间的因果关系。第三,提出了将占用驱动的作用模式 SMIRNA 转化为事件驱动的小分子化学探针(例如 RNA 切割器和降解器)的新兴模式。最后,讨论了小分子 RNA 治疗领域的未来,以及开发有效且选择性的以 RNA 为中心的化学探针需要克服的障碍。











































 京公网安备 11010802027423号
京公网安备 11010802027423号