当前位置:
X-MOL 学术
›
Biopolymers
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Understanding mass spectrometry images: complexity to clarity with machine learning
Biopolymers ( IF 2.9 ) Pub Date : 2020-09-16 , DOI: 10.1002/bip.23400 Wil Gardner 1, 2, 3 , Suzanne M Cutts 2 , Don R Phillips 2 , Paul J Pigram 1
Biopolymers ( IF 2.9 ) Pub Date : 2020-09-16 , DOI: 10.1002/bip.23400 Wil Gardner 1, 2, 3 , Suzanne M Cutts 2 , Don R Phillips 2 , Paul J Pigram 1
Affiliation
|
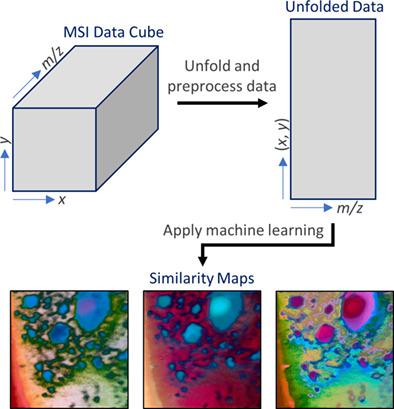
The application of artificial intelligence and machine learning to hyperspectral mass spectrometry imaging (MSI) data has received considerable attention over recent years. Various methodologies have shown great promise in their ability to handle the complexity and size of MSI data sets. Advances in this area have been particularly appealing for MSI of biological samples, which typically produce highly complicated data with often subtle relationships between features. There are many different machine learning approaches that have been applied to MSI data over the past two decades. In this review, we focus on a subset of non-linear machine learning techniques that have mostly only been applied in the past 5 years. Specifically, we review the use of the self-organizing map (SOM), SOM with relational perspective mapping (SOM-RPM), t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP). While not their only functionality, we have grouped these techniques based on their ability to produce what we refer to as similarity maps. Similarity maps are color representations of hyperspectral data, in which spectral similarity between pixels-that is, their distance in high-dimensional space-is represented by relative color similarity. In discussing these techniques, we describe, briefly, their associated algorithms and functionalities, and also outline applications in MSI research with a strong focus on biological sample types. The aim of this review is therefore to introduce this relatively recent paradigm for visualizing and exploring hyperspectral MSI, while also providing a comparison between each technique discussed.
中文翻译:

理解质谱图像:通过机器学习从复杂到清晰
近年来,人工智能和机器学习在高光谱质谱成像 (MSI) 数据中的应用受到了相当多的关注。各种方法在处理 MSI 数据集的复杂性和大小的能力方面显示出了巨大的希望。该领域的进展对生物样本的 MSI 尤其具有吸引力,其通常会产生高度复杂的数据,特征之间通常具有微妙的关系。在过去的二十年中,有许多不同的机器学习方法已应用于 MSI 数据。在这篇评论中,我们关注非线性机器学习技术的一个子集,这些技术大多只在过去 5 年中得到应用。具体来说,我们回顾了自组织映射 (SOM)、带有关系透视映射 (SOM-RPM) 的 SOM 的使用,t 分布随机邻域嵌入 (t-SNE) 和均匀流形近似和投影 (UMAP)。虽然不是它们的唯一功能,但我们根据它们生成我们称之为相似性图的能力对这些技术进行了分组。相似度图是高光谱数据的颜色表示,其中像素之间的光谱相似度——即它们在高维空间中的距离——由相对颜色相似度表示。在讨论这些技术时,我们简要描述了它们相关的算法和功能,并概述了 MSI 研究中的应用,重点关注生物样本类型。因此,本次审查的目的是介绍这种用于可视化和探索高光谱 MSI 的相对较新的范例,同时还提供所讨论的每种技术之间的比较。
更新日期:2020-09-16
中文翻译:

理解质谱图像:通过机器学习从复杂到清晰
近年来,人工智能和机器学习在高光谱质谱成像 (MSI) 数据中的应用受到了相当多的关注。各种方法在处理 MSI 数据集的复杂性和大小的能力方面显示出了巨大的希望。该领域的进展对生物样本的 MSI 尤其具有吸引力,其通常会产生高度复杂的数据,特征之间通常具有微妙的关系。在过去的二十年中,有许多不同的机器学习方法已应用于 MSI 数据。在这篇评论中,我们关注非线性机器学习技术的一个子集,这些技术大多只在过去 5 年中得到应用。具体来说,我们回顾了自组织映射 (SOM)、带有关系透视映射 (SOM-RPM) 的 SOM 的使用,t 分布随机邻域嵌入 (t-SNE) 和均匀流形近似和投影 (UMAP)。虽然不是它们的唯一功能,但我们根据它们生成我们称之为相似性图的能力对这些技术进行了分组。相似度图是高光谱数据的颜色表示,其中像素之间的光谱相似度——即它们在高维空间中的距离——由相对颜色相似度表示。在讨论这些技术时,我们简要描述了它们相关的算法和功能,并概述了 MSI 研究中的应用,重点关注生物样本类型。因此,本次审查的目的是介绍这种用于可视化和探索高光谱 MSI 的相对较新的范例,同时还提供所讨论的每种技术之间的比较。



























 京公网安备 11010802027423号
京公网安备 11010802027423号