iScience ( IF 4.6 ) Pub Date : 2020-09-15 , DOI: 10.1016/j.isci.2020.101565 Hossein Jashnsaz 1 , Zachary R Fox 2, 3, 4 , Jason J Hughes 1 , Guoliang Li 1 , Brian Munsky 4, 5 , Gregor Neuert 1, 6, 7
|
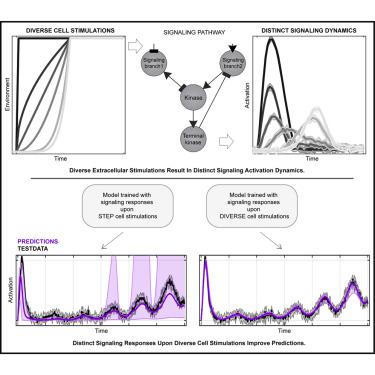
Computationally understanding the molecular mechanisms that give rise to cell signaling responses upon different environmental, chemical, and genetic perturbations is a long-standing challenge that requires models that fit and predict quantitative responses for new biological conditions. Overcoming this challenge depends not only on good models and detailed experimental data but also on the rigorous integration of both. We propose a quantitative framework to perturb and model generic signaling networks using multiple and diverse changing environments (hereafter “kinetic stimulations”) resulting in distinct pathway activation dynamics. We demonstrate that utilizing multiple diverse kinetic stimulations better constrains model parameters and enables predictions of signaling dynamics that would be impossible using traditional dose-response or individual kinetic stimulations. To demonstrate our approach, we use experimentally identified models to predict signaling dynamics in normal, mutated, and drug-treated conditions upon multitudes of kinetic stimulations and quantify which proteins and reaction rates are most sensitive to which extracellular stimulations.
中文翻译:

多种细胞刺激动力学确定预测信号转导模型
通过计算了解在不同环境、化学和遗传扰动下引起细胞信号传导反应的分子机制是一项长期存在的挑战,需要适合和预测新生物条件定量反应的模型。克服这一挑战不仅取决于良好的模型和详细的实验数据,还取决于两者的严格整合。我们提出了一个定量框架,使用多种不同的变化环境(以下称为“动力学刺激”)来扰乱和建模通用信号网络,从而产生不同的通路激活动态。我们证明,利用多种不同的动力学刺激可以更好地限制模型参数,并能够预测信号动力学,而使用传统的剂量反应或单独的动力学刺激是不可能的。为了证明我们的方法,我们使用实验确定的模型来预测正常、突变和药物治疗条件下多种动力学刺激下的信号动力学,并量化哪些蛋白质和反应速率对哪些细胞外刺激最敏感。











































 京公网安备 11010802027423号
京公网安备 11010802027423号