Molecular Cell ( IF 16.0 ) Pub Date : 2020-09-10 , DOI: 10.1016/j.molcel.2020.08.014 Nitin Kapadia 1 , Ziad W El-Hajj 1 , Huan Zheng 1 , Thomas R Beattie 1 , Angela Yu 1 , Rodrigo Reyes-Lamothe 1
|
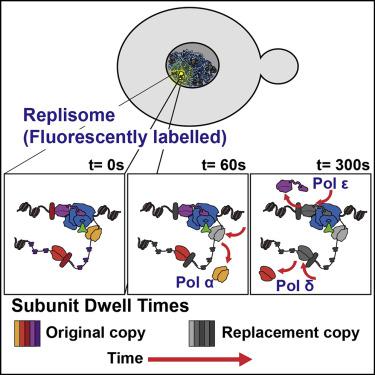
DNA replication is carried out by a multi-protein machine called the replisome. In Saccharomyces cerevisiae, the replisome is composed of over 30 different proteins arranged into multiple subassemblies, each performing distinct activities. Synchrony of these activities is required for efficient replication and preservation of genomic integrity. How this is achieved is particularly puzzling at the lagging strand, where current models of the replisome architecture propose turnover of the canonical lagging strand polymerase, Pol δ, at every cycle of Okazaki fragment synthesis. Here, we established single-molecule fluorescence microscopy protocols to study the binding kinetics of individual replisome subunits in live S. cerevisiae. Our results show long residence times for most subunits at the active replisome, supporting a model where all subassemblies bind tightly and work in a coordinated manner for extended periods, including Pol δ, redefining the architecture of the active eukaryotic replisome.
中文翻译:

真核细胞复制体中复制性DNA聚合酶的加工活性。
DNA复制是通过称为复制体的多蛋白机器进行的。在酿酒酵母中,复制体由30多种不同蛋白质组成,这些蛋白质排列成多个子组件,每个子组件执行不同的活动。这些活动需要同步才能有效地复制和保持基因组完整性。如何做到这一点在滞后链上尤其令人费解,其中复制子体系结构的当前模型提出了在冈崎片段合成的每个循环中规范滞后链聚合酶Polδ的周转率。在这里,我们建立了单分子荧光显微镜协议,以研究活酿酒酵母中单个复制体亚基的结合动力学。。我们的结果表明,大多数亚基在活性复制体上的停留时间较长,从而支持了一个模型,其中所有子组件紧密结合并以协同方式工作了很长时间,包括Polδ,重新定义了活性真核生物复制体的结构。



























 京公网安备 11010802027423号
京公网安备 11010802027423号