当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Comprehensive analysis and ACMG-based classification of CHEK2 variants in hereditary cancer patients.
Human Mutation ( IF 3.3 ) Pub Date : 2020-09-09 , DOI: 10.1002/humu.24110 Gardenia Vargas-Parra 1, 2, 3 , Jesús Del Valle 1, 2, 3 , Paula Rofes 1, 2, 3 , Mireia Gausachs 1, 2 , Agostina Stradella 1, 2, 4 , José M Moreno-Cabrera 1, 2, 3 , Angela Velasco 1, 2, 3 , Eva Tornero 1, 2, 3 , Mireia Menéndez 1, 2, 3 , Xavier Muñoz 1, 2, 3 , Silvia Iglesias 1, 2, 3 , Adriana López-Doriga 5, 6 , Daniel Azuara 1, 2, 3 , Olga Campos 1, 2, 3 , Raquel Cuesta 1, 2, 3 , Esther Darder 1, 2, 3 , Rafael de Cid 7 , Sara González 1, 2, 3 , Alex Teulé 1, 2, 3 , Matilde Navarro 1, 2, 3 , Joan Brunet 1, 2, 3, 8 , Gabriel Capellá 1, 2, 3 , Marta Pineda 1, 2, 3 , Lídia Feliubadaló 1, 2, 3 , Conxi Lázaro 1, 2, 3
Human Mutation ( IF 3.3 ) Pub Date : 2020-09-09 , DOI: 10.1002/humu.24110 Gardenia Vargas-Parra 1, 2, 3 , Jesús Del Valle 1, 2, 3 , Paula Rofes 1, 2, 3 , Mireia Gausachs 1, 2 , Agostina Stradella 1, 2, 4 , José M Moreno-Cabrera 1, 2, 3 , Angela Velasco 1, 2, 3 , Eva Tornero 1, 2, 3 , Mireia Menéndez 1, 2, 3 , Xavier Muñoz 1, 2, 3 , Silvia Iglesias 1, 2, 3 , Adriana López-Doriga 5, 6 , Daniel Azuara 1, 2, 3 , Olga Campos 1, 2, 3 , Raquel Cuesta 1, 2, 3 , Esther Darder 1, 2, 3 , Rafael de Cid 7 , Sara González 1, 2, 3 , Alex Teulé 1, 2, 3 , Matilde Navarro 1, 2, 3 , Joan Brunet 1, 2, 3, 8 , Gabriel Capellá 1, 2, 3 , Marta Pineda 1, 2, 3 , Lídia Feliubadaló 1, 2, 3 , Conxi Lázaro 1, 2, 3
Affiliation
|
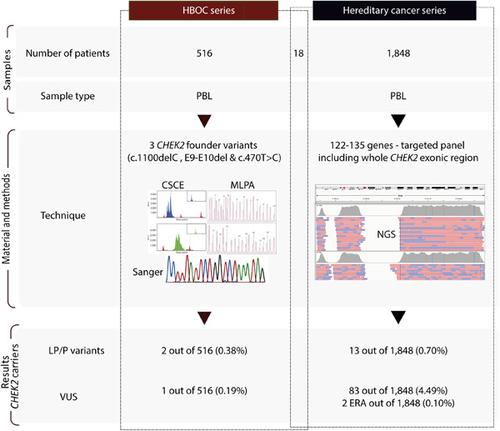
CHEK2 variants are associated with intermediate breast cancer risk, among other cancers. We aimed to comprehensively describe CHEK2 variants in a Spanish hereditary cancer (HC) cohort and adjust the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG‐AMP) guidelines for their classification. First, three CHEK2 frequent variants were screened in a retrospective Hereditary Breast and Ovarian Cancer cohort of 516 patients. After, the whole CHEK2 coding region was analyzed by next‐generation sequencing in 1848 prospective patients with HC suspicion. We refined ACMG‐AMP criteria and applied different combined rules to classify CHEK2 variants and define risk alleles. We identified 10 CHEK2 null variants, 6 missense variants with discordant interpretation in ClinVar database, and 35 additional variants of unknown significance. Twelve variants were classified as (likely)‐pathogenic; two can also be considered “established risk‐alleles” and one as “likely risk‐allele.” The prevalence of (likely)‐pathogenic variants in the HC cohort was 0.8% (1.3% in breast cancer patients and 1.0% in hereditary nonpolyposis colorectal cancer patients). Here, we provide ACMG adjustment guidelines to classify CHEK2 variants. We hope that this study would be useful for variant classification of other genes with low effect variants.
中文翻译:

遗传性癌症患者 CHEK2 变异的综合分析和基于 ACMG 的分类。
CHEK2变体与中等乳腺癌风险相关,以及其他癌症。我们旨在全面描述西班牙遗传性癌症 (HC) 队列中的CHEK2变异,并调整美国医学遗传学和基因组学学院和分子病理学协会 (ACMG-AMP) 的分类指南。首先,在 516 名患者的回顾性遗传性乳腺癌和卵巢癌队列中筛选了三个CHEK2频繁变异。之后,通过二代测序分析了 1848 名疑似 HC 的前瞻性患者的整个CHEK2编码区。我们改进了 ACMG-AMP 标准并应用不同的组合规则对CHEK2进行分类变体并定义风险等位基因。我们确定了 10 个CHEK2空变体、6 个在 ClinVar 数据库中解释不一致的错义变体和 35 个意义不明的其他变体。12 种变异被归类为(可能的)致病性;两个也可以被认为是“既定的风险等位基因”,一个是“可能的风险等位基因”。HC 队列中(可能的)致病变异的患病率为 0.8%(乳腺癌患者为 1.3%,遗传性非息肉病性结直肠癌患者为 1.0%)。在这里,我们提供了 ACMG 调整指南来对CHEK2变体进行分类。我们希望这项研究对其他低效应变异基因的变异分类有用。
更新日期:2020-09-09
中文翻译:

遗传性癌症患者 CHEK2 变异的综合分析和基于 ACMG 的分类。
CHEK2变体与中等乳腺癌风险相关,以及其他癌症。我们旨在全面描述西班牙遗传性癌症 (HC) 队列中的CHEK2变异,并调整美国医学遗传学和基因组学学院和分子病理学协会 (ACMG-AMP) 的分类指南。首先,在 516 名患者的回顾性遗传性乳腺癌和卵巢癌队列中筛选了三个CHEK2频繁变异。之后,通过二代测序分析了 1848 名疑似 HC 的前瞻性患者的整个CHEK2编码区。我们改进了 ACMG-AMP 标准并应用不同的组合规则对CHEK2进行分类变体并定义风险等位基因。我们确定了 10 个CHEK2空变体、6 个在 ClinVar 数据库中解释不一致的错义变体和 35 个意义不明的其他变体。12 种变异被归类为(可能的)致病性;两个也可以被认为是“既定的风险等位基因”,一个是“可能的风险等位基因”。HC 队列中(可能的)致病变异的患病率为 0.8%(乳腺癌患者为 1.3%,遗传性非息肉病性结直肠癌患者为 1.0%)。在这里,我们提供了 ACMG 调整指南来对CHEK2变体进行分类。我们希望这项研究对其他低效应变异基因的变异分类有用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号