当前位置:
X-MOL 学术
›
Hum. Mutat.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Frequency spectrum of rare and clinically relevant markers in multiethnic Indian populations (ClinIndb): A resource for genomic medicine in India.
Human Mutation ( IF 3.3 ) Pub Date : 2020-09-09 , DOI: 10.1002/humu.24102 Ankita Narang 1 , Bharathram Uppilli 1, 2 , Asokachandran Vivekanand 1, 2 , Salwa Naushin 1, 2 , Arti Yadav 3 , Khushboo Singhal 1, 2 , Uzma Shamim 1 , Pooja Sharma 1, 2 , Sana Zahra 1, 2 , Aradhana Mathur 1 , Malika Seth 1 , Shaista Parveen 1 , Archana Vats 1 , Sara Hillman 4 , Padma Dolma 5 , Binuja Varma 3 , Vandana Jain 6 , , Bhavana Prasher 1, 2, 3 , Shantanu Sengupta 1, 2 , Mitali Mukerji 1, 2, 3 , Mohammed Faruq 1, 2
Human Mutation ( IF 3.3 ) Pub Date : 2020-09-09 , DOI: 10.1002/humu.24102 Ankita Narang 1 , Bharathram Uppilli 1, 2 , Asokachandran Vivekanand 1, 2 , Salwa Naushin 1, 2 , Arti Yadav 3 , Khushboo Singhal 1, 2 , Uzma Shamim 1 , Pooja Sharma 1, 2 , Sana Zahra 1, 2 , Aradhana Mathur 1 , Malika Seth 1 , Shaista Parveen 1 , Archana Vats 1 , Sara Hillman 4 , Padma Dolma 5 , Binuja Varma 3 , Vandana Jain 6 , , Bhavana Prasher 1, 2, 3 , Shantanu Sengupta 1, 2 , Mitali Mukerji 1, 2, 3 , Mohammed Faruq 1, 2
Affiliation
|
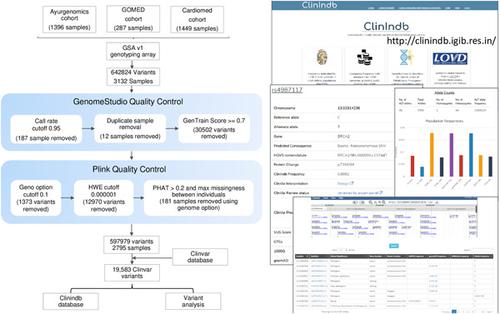
There have been concerted efforts toward cataloging rare and deleterious variants in different world populations using high‐throughput genotyping and sequencing‐based methods. The Indian population is underrepresented or its information with respect to clinically relevant variants is sparse in public data sets. The aim of this study was to estimate the burden of monogenic disease‐causing variants in Indian populations. Toward this, we have assessed the frequency profile of monogenic phenotype‐associated ClinVar variants. The study utilized a genotype data set (global screening array, Illumina) from 2795 individuals (multiple in‐house genomics cohorts) representing diverse ethnic and geographically distinct Indian populations. Of the analyzed variants from Global Screening Array, ~9% were found to be informative and were either not known earlier or underrepresented in public databases in terms of their frequencies. These variants were linked to disorders, namely inborn errors of metabolism, monogenic diabetes, hereditary cancers, and various other hereditary conditions. We have also shown that our study cohort is genetically a better representative of the Indian population than its representation in the 1000 Genome Project (South Asians). We have created a database, ClinIndb, linked to the Leiden Open Variation Database, to help clinicians and researchers in diagnosis, counseling, and development of appropriate genetic screening tools relevant to the Indian populations and Indians living abroad.
中文翻译:

印度多民族人群中罕见和临床相关标志物的频谱 (ClinIndb):印度基因组医学资源。
使用高通量基因分型和基于测序的方法对不同世界人群中的稀有和有害变异进行编目已经做出了一致的努力。印度人口代表性不足,或者其关于临床相关变异的信息在公共数据集中稀少。本研究的目的是估计印度人群中引起单基因疾病的变异的负担。为此,我们评估了单基因表型相关 ClinVar 变异的频率分布。该研究使用了来自 2795 个人(多个内部基因组队列)的基因型数据集(全球筛选阵列,Illumina),代表不同种族和地理上不同的印度人口。在来自 Global Screening Array 的分析变体中,约 9% 被发现是信息丰富的,并且在公共数据库中的频率方面要么不早知道要么代表性不足。这些变异与疾病有关,即先天性代谢缺陷、单基因糖尿病、遗传性癌症和各种其他遗传性疾病。我们还表明,与 1000 基因组计划(南亚人)相比,我们的研究队列在遗传上更能代表印度人口。我们创建了一个与莱顿开放变异数据库相关联的数据库 ClinIndb,以帮助临床医生和研究人员诊断、咨询和开发与印度人口和居住在国外的印度人相关的适当基因筛查工具。即先天性代谢缺陷、单基因糖尿病、遗传性癌症和其他各种遗传性疾病。我们还表明,与 1000 基因组计划(南亚人)相比,我们的研究队列在遗传上更能代表印度人口。我们创建了一个与莱顿开放变异数据库相关联的数据库 ClinIndb,以帮助临床医生和研究人员诊断、咨询和开发与印度人口和居住在国外的印度人相关的适当基因筛查工具。即先天性代谢缺陷、单基因糖尿病、遗传性癌症和其他各种遗传性疾病。我们还表明,与 1000 基因组计划(南亚人)相比,我们的研究队列在遗传上更能代表印度人口。我们创建了一个数据库 ClinIndb,与莱顿开放变异数据库相关联,以帮助临床医生和研究人员进行诊断、咨询和开发与印度人口和居住在国外的印度人相关的适当基因筛查工具。
更新日期:2020-10-30
中文翻译:

印度多民族人群中罕见和临床相关标志物的频谱 (ClinIndb):印度基因组医学资源。
使用高通量基因分型和基于测序的方法对不同世界人群中的稀有和有害变异进行编目已经做出了一致的努力。印度人口代表性不足,或者其关于临床相关变异的信息在公共数据集中稀少。本研究的目的是估计印度人群中引起单基因疾病的变异的负担。为此,我们评估了单基因表型相关 ClinVar 变异的频率分布。该研究使用了来自 2795 个人(多个内部基因组队列)的基因型数据集(全球筛选阵列,Illumina),代表不同种族和地理上不同的印度人口。在来自 Global Screening Array 的分析变体中,约 9% 被发现是信息丰富的,并且在公共数据库中的频率方面要么不早知道要么代表性不足。这些变异与疾病有关,即先天性代谢缺陷、单基因糖尿病、遗传性癌症和各种其他遗传性疾病。我们还表明,与 1000 基因组计划(南亚人)相比,我们的研究队列在遗传上更能代表印度人口。我们创建了一个与莱顿开放变异数据库相关联的数据库 ClinIndb,以帮助临床医生和研究人员诊断、咨询和开发与印度人口和居住在国外的印度人相关的适当基因筛查工具。即先天性代谢缺陷、单基因糖尿病、遗传性癌症和其他各种遗传性疾病。我们还表明,与 1000 基因组计划(南亚人)相比,我们的研究队列在遗传上更能代表印度人口。我们创建了一个与莱顿开放变异数据库相关联的数据库 ClinIndb,以帮助临床医生和研究人员诊断、咨询和开发与印度人口和居住在国外的印度人相关的适当基因筛查工具。即先天性代谢缺陷、单基因糖尿病、遗传性癌症和其他各种遗传性疾病。我们还表明,与 1000 基因组计划(南亚人)相比,我们的研究队列在遗传上更能代表印度人口。我们创建了一个数据库 ClinIndb,与莱顿开放变异数据库相关联,以帮助临床医生和研究人员进行诊断、咨询和开发与印度人口和居住在国外的印度人相关的适当基因筛查工具。











































 京公网安备 11010802027423号
京公网安备 11010802027423号