Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2020-09-05 , DOI: 10.1016/j.jmb.2020.08.024 Freda E-C Jen 1 , Adeana L Scott 1 , Aimee Tan 1 , Kate L Seib 1 , Michael P Jennings 1
|
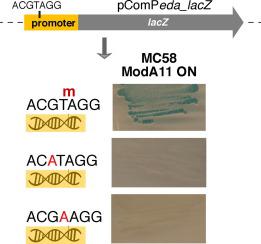
Phase-variable DNA methyltransferases (Mods) mediate epigenetic regulation of gene expression. These phase-variable regulons, called phasevarions, have been shown to regulate virulence and immunoevasion in multiple bacterial pathogens. How genome methylation switching mediates gene regulation is unresolved. Neisseria meningitidis remains a major cause of sepsis and meningitis worldwide. Previously, we reported that phase variation (rapid on/off switching) of the meningococcal ModA11 methyltransferase regulates 285 genes. Here we show a bioinformatic analysis that reveals only 26 of the regulated genes have a methylation site located upstream of the gene with potential for direct effect of methylation on transcription. To investigate how methylation changes are “read” to alter gene expression, we used a lacZ gene fusion approach. We showed a 182-nucleotide region upstream of the eda gene (Entner–Doudoroff aldolase) is sufficient to impart methylation-dependent regulation of eda. Site-directed mutagenesis of the 5′-ACGTm6AGG-3′ ModA11 site upstream of the eda gene showed that methylation of this site modulates eda expression. We show that eda is regulated by the PhoB homolog MisR, and that a MisR binding motif overlaps with the ModA11 methylation site. In a MisR mutant, regulation of eda is uncoupled from regulation by ModA11 phasevarion switching. The on/off switching of ModA11 leads to the presence or absence of a N6-methyladenine modification at thousands of sites in the genome. Most of these modifications have no impact on gene regulation. Moreover, the majority of the 285 gene regulon that is controlled by ModA11 phasevarion switching (259/285) are not directly controlled by methylation changes in the promoter region of the regulated genes. Our data are consistent with direct control via methylation of a subset of the regulon, like Eda, whose regulation will trigger secondary effects in expression of many genes.
中文翻译:

脑膜炎奈瑟氏球菌的ModA11 III型DNA甲基转移酶的随机转换通过eda启动子区域的甲基化变化来调节Enter-Doudoroff醛缩酶的表达。
相变DNA甲基转移酶(Mods)介导基因表达的表观遗传调控。这些被称为相变的相变调节剂已被证明可调节多种细菌病原体的毒力和免疫逃逸。基因组甲基化开关如何介导基因调控尚未解决。脑膜炎奈瑟氏球菌仍然是全世界败血症和脑膜炎的主要原因。以前,我们报道了脑膜炎球菌ModA11甲基转移酶的相变(快速开关)调节285个基因。在这里,我们显示了一种生物信息学分析,该分析揭示了仅26个受调控基因的甲基化位点位于该基因的上游,具有甲基化对转录的直接作用的潜力。为了研究如何“读取”甲基化变化以改变基因表达,我们使用了lacZ基因融合方法。我们显示eda基因上游的182个核苷酸区域(Entner-Doudoroff aldolase)足以赋予eda甲基化依赖性调控。eda基因上游5'-ACGT m6 AGG-3'ModA11位点的定点诱变显示该位点的甲基化调节eda表达。我们显示eda受PhoB同源MisR调控,并且MisR结合基序与ModA11甲基化位点重叠。在MisR突变体中,对eda的调控通过ModA11相变开关与调节解耦。ModA11的开/关切换会导致基因组中数千个位点存在或不存在N6-甲基腺嘌呤修饰。这些修饰大多数对基因调控没有影响。此外,由ModA11相变切换(259/285)控制的大多数285基因调节子不受调节基因启动子区域的甲基化变化直接控制。我们的数据与通过调节子区域(如Eda)的甲基化直接控制一致,其调控将触发许多基因表达的次生效应。











































 京公网安备 11010802027423号
京公网安备 11010802027423号