Journal of Proteomics ( IF 2.8 ) Pub Date : 2020-09-03 , DOI: 10.1016/j.jprot.2020.103965 Bing Wang 1 , Junhui Hao 1 , Ni Pan 1 , Zhiwei Wang 1 , Yinxuan Chen 1 , Cuihong Wan 1
|
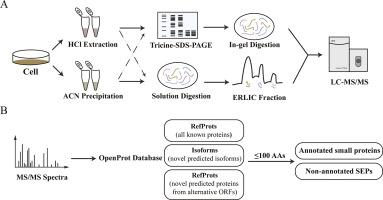
The small proteins and short open reading frames encoded peptides (SEPs) are of fundamental importance because of their essential roles in biological processes. However, the annotation or identification of them is challenging, in part owing to the limitation of the traditional genome annotation pipeline and their inherent characteristics of low abundance and low molecular weight. To discover and characterize SEPs in Hep3B cell line, we developed an optimized peptidomic assay by combining different peptide extraction and separation methods. The organic solvent precipitation method in peptidomic showed promotion in the enrichment of low molecular proteins or peptides, and the data clearly showed a beneficial effect from the reduction of sample complexity, resulting in high-quality MS/MS spectra. Furthermore, different strategies exhibited good complementarity in improving the total amount of small proteins and their sequence coverage. In total, 1192 proteins within less than 100 amino acids were identified, including 271 newly discovered SEPs that been annotated in the OpenProt database and 147 SEPs of them encoded from ncRNA or lincRNA. Results in this work provide robust evidence to date that the human proteome is more complicated than previously appreciated, and this will be a benefit to discoveries of proteins without function annotation.
Significance
In this work, methods were optimized to identify SEPs in Hep3B. The organic solvent precipitation presents promotion in enrichment of low molecular proteins or peptides, and the data clearly showed a beneficial effect from the reduction of sample complexity, resulting in high quality MS/MS spectra. Different strategies exhibited good complementarity in improving total amount of small proteins and their sequence coverage. In total, 1192 proteins within less than 100 amino acids were identified, including 271 newly discovered SEPs that been annotated in the OpenProt database and 147 SEPs of them encoded from ncRNA or lincRNA. Furthermore, 22 SEPs generated from the uORF may has potential effect in translation control, and 149 newly identified SEPs have known functional domains or cross-species conservation. Results in this work present robust evidence for the coding potential of the ignored region of human genomes and may provide additional insights into tumor biology.
中文翻译:

鉴定和分析Hep3B细胞中的小蛋白质和短开放阅读框编码的肽。
小蛋白质和短开放阅读框编码的肽(SEP)具有根本的重要性,因为它们在生物学过程中起着至关重要的作用。然而,对它们的注释或鉴定具有挑战性,部分原因是传统基因组注释管道的局限性及其低丰度和低分子量的固有特征。为了发现和表征Hep3B细胞系中的SEP,我们通过结合不同的肽提取和分离方法开发了一种优化的肽组测定法。肽组学中的有机溶剂沉淀法显示出促进了低分子蛋白质或肽的富集,并且数据清楚地表明了降低样品复杂性的有益效果,从而获得了高质量的MS / MS谱图。此外,不同的策略在改善小蛋白质的总量及其序列覆盖率方面显示出良好的互补性。总共鉴定出不足100个氨基酸的1192种蛋白质,其中包括271种新发现的SEP(已在OpenProt数据库中注释),其中147种SEP由ncRNA或lincRNA编码。迄今为止,这项工作的结果提供了有力的证据,证明人类蛋白质组比以前认为的更为复杂,这对发现没有功能注释的蛋白质将是有益的。
意义
在这项工作中,方法被优化以识别Hep3B中的SEP。有机溶剂的沉淀促进了低分子蛋白质或肽的富集,并且数据清楚地表明了降低样品复杂性的有益效果,从而获得了高质量的MS / MS谱图。不同的策略在提高小蛋白的总量及其序列覆盖率方面表现出良好的互补性。总共鉴定出不足100个氨基酸的1192种蛋白质,其中包括271种新发现的SEP(已在OpenProt数据库中注释),其中147种SEP由ncRNA或lincRNA编码。此外,从uORF生成的22个SEP可能在翻译控制中具有潜在作用,而149个新发现的SEP具有已知的功能域或跨物种保守性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号