当前位置:
X-MOL 学术
›
J. Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
XGen: Real-Space Fitting of Complex Ligand Conformational Ensembles to X-ray Electron Density Maps.
Journal of Medicinal Chemistry ( IF 6.8 ) Pub Date : 2020-09-02 , DOI: 10.1021/acs.jmedchem.0c01373 Ajay N Jain 1 , Ann E Cleves 2 , Alexander C Brueckner 3 , Charles A Lesburg 3 , Qiaolin Deng 3 , Edward C Sherer 3 , Mikhail Y Reibarkh 3
Journal of Medicinal Chemistry ( IF 6.8 ) Pub Date : 2020-09-02 , DOI: 10.1021/acs.jmedchem.0c01373 Ajay N Jain 1 , Ann E Cleves 2 , Alexander C Brueckner 3 , Charles A Lesburg 3 , Qiaolin Deng 3 , Edward C Sherer 3 , Mikhail Y Reibarkh 3
Affiliation
|
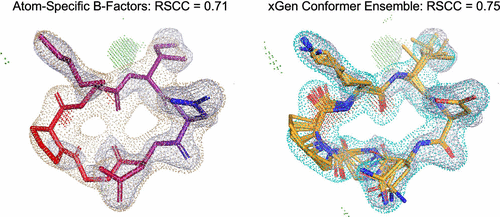
We report a new method for X-ray density ligand fitting and refinement that is suitable for a wide variety of small-molecule ligands, including macrocycles. The approach (called “xGen”) augments a force field energy calculation with an electron density fitting restraint that yields an energy reward during the restrained conformational search. The resulting conformer pools balance goodness-of-fit with ligand strain. Real-space refinement from pre-existing ligand coordinates of 150 macrocycles resulted in occupancy-weighted conformational ensembles that exhibited low strain energy. The xGen ensembles improved upon electron density fit compared with the PDB reference coordinates without making use of atom-specific B-factors. Similarly, on nonmacrocycles, de novo fitting produced occupancy-weighted ensembles of many conformers that were generally better-quality density fits than the deposited primary/alternate conformational pairs. The results suggest ubiquitous low-energy ligand conformational ensembles in X-ray diffraction data and provide an alternative to using B-factors as model parameters.
中文翻译:

XGen:复杂配体构象集合对X射线电子密度图的真实空间拟合。
我们报告了一种适用于各种广泛的小分子配体(包括大环)的X射线密度配体拟合和提纯的新方法。该方法(称为“ xGen”)通过电子密度拟合约束来增强力场能量计算,该约束在约束构象搜索过程中产生能量奖励。最终形成的构象库在拟合优度和配体应变之间达到平衡。从150个大环的预先存在的配体坐标进行的实际空间细化导致占用权重的构象集合表现出较低的应变能。与PDB参考坐标相比,在不使用原子特定的B因子的情况下,通过电子密度拟合,xGen集成得到了改善。同样地,在非宏观自行车上,从头开始拟合产生的许多构象的占用加权合奏,通常比沉积的初级/交替构象对具有更好的质量密度拟合。结果表明,X射线衍射数据中普遍存在低能配体构象集合,这为使用B因子作为模型参数提供了一种替代方法。
更新日期:2020-09-24
中文翻译:

XGen:复杂配体构象集合对X射线电子密度图的真实空间拟合。
我们报告了一种适用于各种广泛的小分子配体(包括大环)的X射线密度配体拟合和提纯的新方法。该方法(称为“ xGen”)通过电子密度拟合约束来增强力场能量计算,该约束在约束构象搜索过程中产生能量奖励。最终形成的构象库在拟合优度和配体应变之间达到平衡。从150个大环的预先存在的配体坐标进行的实际空间细化导致占用权重的构象集合表现出较低的应变能。与PDB参考坐标相比,在不使用原子特定的B因子的情况下,通过电子密度拟合,xGen集成得到了改善。同样地,在非宏观自行车上,从头开始拟合产生的许多构象的占用加权合奏,通常比沉积的初级/交替构象对具有更好的质量密度拟合。结果表明,X射线衍射数据中普遍存在低能配体构象集合,这为使用B因子作为模型参数提供了一种替代方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号