Water Research ( IF 12.8 ) Pub Date : 2020-09-01 , DOI: 10.1016/j.watres.2020.116372 Michael Lukumbuzya 1 , Jannie Munk Kristensen 2 , Katharina Kitzinger 3 , Andreas Pommerening-Röser 4 , Per Halkjær Nielsen 2 , Michael Wagner 5 , Holger Daims 6 , Petra Pjevac 7
|
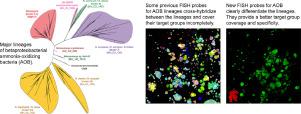
Ammonia-oxidizing bacteria (AOB) of the betaproteobacterial genera Nitrosomonas and Nitrosospira are key nitrifying microorganisms in many natural and engineered ecosystems. Since many AOB remain uncultured, fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes has been one of the most widely used approaches to study the community composition, abundance, and other features of AOB directly in environmental samples. However, the established and widely used AOB-specific 16S rRNA-targeted FISH probes were designed up to two decades ago, based on much smaller rRNA gene sequence datasets than available today. Several of these probes cover their target AOB lineages incompletely and suffer from a weak target specificity, which causes cross-hybridization of probes that should detect different AOB lineages. Here, a set of new highly specific 16S rRNA-targeted oligonucleotide probes was developed and experimentally evaluated that complements the existing probes and enables the specific detection and differentiation of the known, major phylogenetic clusters of betaproteobacterial AOB. The new probes were successfully applied to visualize and quantify AOB in activated sludge and biofilm samples from seven pilot- and full-scale wastewater treatment systems. Based on its improved target group coverage and specificity, the refined probe set will facilitate future in situ analyses of AOB.
中文翻译:

一套精致的rRNA靶向寡核苷酸探针,用于原位检测和定量氨氧化细菌。
β变形杆菌属亚硝化单胞菌和亚硝基螺菌属的氨氧化细菌(AOB)是许多自然和工程生态系统中的关键硝化微生物。由于许多AOB仍未培养,因此以rRNA靶向的寡核苷酸探针进行荧光原位杂交(FISH)已成为直接研究环境样品中AOB的群落组成,丰度和其他特征的最广泛使用的方法之一。然而,已建立且广泛使用的针对AOB的16S rRNA靶向FISH探针是在二十年前设计的,基于比今天更小的rRNA基因序列数据集。这些探针中的几种探针未完全覆盖其靶AOB谱系,并且靶标特异性较弱,这会导致应检测不同AOB谱系的探针交叉杂交。这里,研发了一组新的高度特异性的16S rRNA靶向寡核苷酸探针,并进行了实验评估,该探针可补充现有探针,并能够特异性检测和区分已知的,主要的β变形细菌AOB系统发生簇。新探针已成功应用于可视化和量化来自七个中试和大型废水处理系统的活性污泥和生物膜样品中的AOB。基于改进的目标组覆盖范围和特异性,改进的探针组将有助于将来对AOB进行原位分析。新探针已成功应用于可视化和量化来自七个中试和大型废水处理系统的活性污泥和生物膜样品中的AOB。基于改进的目标组覆盖范围和特异性,改进的探针组将有助于将来对AOB进行原位分析。新探针已成功应用于可视化和量化来自七个中试和大型废水处理系统的活性污泥和生物膜样品中的AOB。基于改进的目标群覆盖范围和特异性,改进的探针组将有助于将来对AOB进行原位分析。



























 京公网安备 11010802027423号
京公网安备 11010802027423号