Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2020-09-02 , DOI: 10.1016/j.csbj.2020.08.023 Kai Zheng 1 , Zhu-Hong You 2 , Lei Wang 2, 3 , Zhen-Hao Guo 2
|
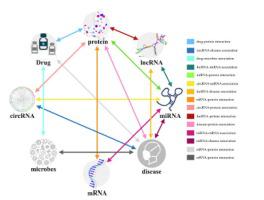
Benefiting from advances in high-throughput experimental techniques, important regulatory roles of miRNAs, lncRNAs, and proteins, as well as biological property information, are gradually being complemented. As the key data support to promote biomedical research, domain knowledge such as intermolecular relationships that are increasingly revealed by molecular genome-wide analysis is often used to guide the discovery of potential associations. However, the method of performing network representation learning from the perspective of the global biological network is scarce. These methods cover a very limited type of molecular associations and are therefore not suitable for more comprehensive analysis of molecular network representation information. In this study, we propose a computational model based on the Biological network for predicting potential associations between miRNAs and diseases called iMDA-BN. The iMDA-BN has three significant advantages: I) It uses a new method to describe disease and miRNA characteristics which analyzes node representation information for disease and miRNA from the perspective of biological networks. II) It can predict unproven associations even if miRNAs and diseases do not appear in the biological network. III) Accurate description of miRNA characteristics from biological properties based on high-throughput sequence information. The iMDA-BN predictor achieves an AUC of 0.9145 and an accuracy of 84.49% on the miRNA-disease association baseline dataset, and it can also achieve an AUC of 0.8765 and an accuracy of 80.96% when predicting unknown diseases and miRNAs in the biological network. Compared to existing miRNA-disease association prediction methods, iMDA-BN has higher accuracy and the advantage of predicting unknown associations. In addition, 45, 49, and 49 of the top 50 miRNA-disease associations with the highest predicted scores were confirmed in the case studies, respectively.
中文翻译:

iMDA-BN:基于生物网络和图嵌入算法的 miRNA 疾病关联识别
受益于高通量实验技术的进步,miRNA、lncRNA和蛋白质的重要调控作用以及生物学特性信息正在逐渐得到补充。作为促进生物医学研究的关键数据支撑,分子全基因组分析日益揭示的分子间关系等领域知识常常被用来指导潜在关联的发现。然而,从全局生物网络的角度进行网络表示学习的方法却很少。这些方法涵盖了非常有限的分子关联类型,因此不适合对分子网络表示信息进行更全面的分析。在这项研究中,我们提出了一种基于生物网络的计算模型,用于预测 miRNA 与疾病之间的潜在关联,称为 iMDA-BN。 iMDA-BN具有三个显着优点:一)它采用了一种新的方法来描述疾病和miRNA特征,从生物网络的角度分析疾病和miRNA的节点表示信息。 II) 即使 miRNA 和疾病没有出现在生物网络中,它也可以预测未经证实的关联。 III) 基于高通量序列信息从生物学特性准确描述miRNA特征。 iMDA-BN预测器在miRNA-疾病关联基线数据集上实现了0.9145的AUC和84.49%的准确率,并且在预测生物网络中的未知疾病和miRNA时也可以实现0.8765的AUC和80.96%的准确率。与现有的miRNA-疾病关联预测方法相比,iMDA-BN具有更高的准确性和预测未知关联的优势。 此外,预测得分最高的前 50 个 miRNA 与疾病关联中,分别有 45、49 和 49 个在案例研究中得到了证实。











































 京公网安备 11010802027423号
京公网安备 11010802027423号