Cell Metabolism ( IF 27.7 ) Pub Date : 2020-09-01 , DOI: 10.1016/j.cmet.2020.07.017 Hana Antonicka 1 , Zhen-Yuan Lin 2 , Alexandre Janer 1 , Mari J Aaltonen 1 , Woranontee Weraarpachai 3 , Anne-Claude Gingras 4 , Eric A Shoubridge 1
|
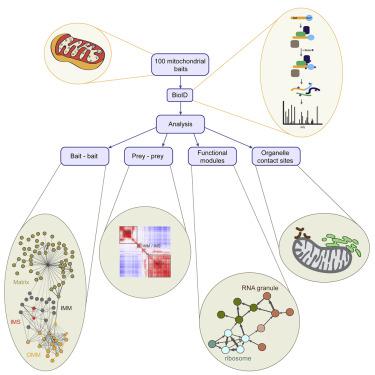
We used BioID, a proximity-dependent biotinylation assay with 100 mitochondrial baits from all mitochondrial sub-compartments, to create a high-resolution human mitochondrial proximity interaction network. We identified 1,465 proteins, producing 15,626 unique high-confidence proximity interactions. Of these, 528 proteins were previously annotated as mitochondrial, nearly half of the mitochondrial proteome defined by Mitocarta 2.0. Bait-bait analysis showed a clear separation of mitochondrial compartments, and correlation analysis among preys across all baits allowed us to identify functional clusters involved in diverse mitochondrial functions and to assign uncharacterized proteins to specific modules. We demonstrate that this analysis can assign isoforms of the same mitochondrial protein to different mitochondrial sub-compartments and show that some proteins may have multiple cellular locations. Outer membrane baits showed specific proximity interactions with cytosolic proteins and proteins in other organellar membranes, suggesting specialization of proteins responsible for contact site formation between mitochondria and individual organelles.
中文翻译:

高密度人类线粒体邻近相互作用网络。
我们使用 BioID,一种接近依赖性生物素化测定,具有来自所有线粒体子隔室的 100 个线粒体诱饵,以创建高分辨率的人类线粒体接近相互作用网络。我们鉴定了 1,465 种蛋白质,产生了 15,626 种独特的高置信度邻近相互作用。其中,528 种蛋白质之前被注释为线粒体,几乎是 Mitocarta 2.0 定义的线粒体蛋白质组的一半。诱饵分析显示线粒体隔室的清晰分离,所有诱饵的猎物之间的相关性分析使我们能够识别涉及不同线粒体功能的功能簇,并将未表征的蛋白质分配给特定模块。我们证明该分析可以将相同线粒体蛋白质的同种型分配给不同的线粒体亚室,并表明某些蛋白质可能具有多个细胞位置。外膜诱饵显示出与细胞质蛋白和其他细胞器膜中的蛋白质的特定邻近相互作用,表明负责线粒体和单个细胞器之间接触位点形成的蛋白质的特化。











































 京公网安备 11010802027423号
京公网安备 11010802027423号