Cell Systems ( IF 9.0 ) Pub Date : 2020-08-31 , DOI: 10.1016/j.cels.2020.08.003 Payam Dibaeinia 1 , Saurabh Sinha 2
|
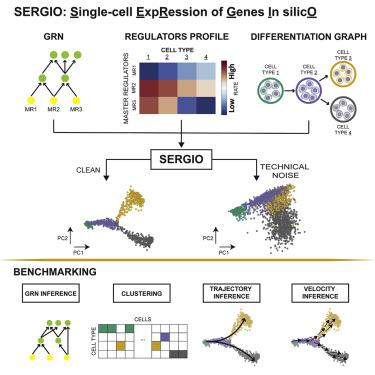
A common approach to benchmarking of single-cell transcriptomics tools is to generate synthetic datasets that statistically resemble experimental data. However, most existing single-cell simulators do not incorporate transcription factor-gene regulatory interactions that underlie expression dynamics. Here, we present SERGIO, a simulator of single-cell gene expression data that models the stochastic nature of transcription as well as regulation of genes by multiple transcription factors according to a user-provided gene regulatory network. SERGIO can simulate any number of cell types in steady state or cells differentiating to multiple fates. We show that datasets generated by SERGIO are statistically comparable to experimental data generated by Illumina HiSeq2000, Drop-seq, Illumina 10X chromium, and Smart-seq. We use SERGIO to benchmark several single-cell analysis tools, including GRN inference methods, and identify Tcf7, Gata3, and Bcl11b as key drivers of T cell differentiation by performing in silico knockout experiments. SERGIO is freely available for download here: https://github.com/PayamDiba/SERGIO.
中文翻译:

SERGIO:基因调控网络引导的单细胞表达模拟器。
单细胞转录组学工具基准测试的常见方法是生成统计上类似于实验数据的合成数据集。然而,大多数现有的单细胞模拟器并未纳入表达动力学基础的转录因子-基因调控相互作用。在这里,我们推出了 SERGIO,这是一个单细胞基因表达数据的模拟器,它可以根据用户提供的基因调控网络对转录的随机性质以及多个转录因子对基因的调控进行建模。 SERGIO 可以模拟任意数量的稳态细胞类型或分化为多种命运的细胞。我们表明,SERGIO 生成的数据集在统计上与 Illumina HiSeq2000、Drop-seq、Illumina 10X chromium 和 Smart-seq 生成的实验数据相当。我们使用 SERGIO 对多种单细胞分析工具(包括 GRN 推断方法)进行基准测试,并通过进行计算机敲除实验,确定 Tcf7、Gata3 和 Bcl11b 作为 T 细胞分化的关键驱动因素。 SERGIO 可在此处免费下载:https://github.com/PayamDiba/SERGIO。









































 京公网安备 11010802027423号
京公网安备 11010802027423号