Structure ( IF 5.7 ) Pub Date : 2020-08-27 , DOI: 10.1016/j.str.2020.08.005 Oleg V Sobolev 1 , Pavel V Afonine 1 , Nigel W Moriarty 1 , Maarten L Hekkelman 2 , Robbie P Joosten 2 , Anastassis Perrakis 2 , Paul D Adams 3
|
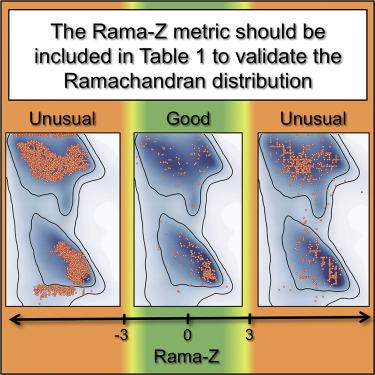
Ramachandran plots report the distribution of the (ϕ, ψ) torsion angles of the protein backbone and are one of the best quality metrics of experimental structure models. Typically, validation software reports the number of residues belonging to “outlier,” “allowed,” and “favored” regions. While “zero unexplained outliers” can be considered the current “gold standard,” this can be misleading if deviations from expected distributions are not considered. We revisited the Ramachandran Z score (Rama-Z), a quality metric introduced more than two decades ago but underutilized. We describe a reimplementation of the Rama-Z score in the Computational Crystallography Toolbox along with an algorithm to estimate its uncertainty for individual models; final implementations are available in Phenix and PDB-REDO. We discuss the interpretation of the Rama-Z score and advocate including it in the validation reports provided by the Protein Data Bank. We also advocate reporting it alongside the outlier/allowed/favored counts in structural publications.
中文翻译:

全球 Ramachandran 分数可识别具有不太可能立体化学的蛋白质结构。
Ramachandran 图报告了蛋白质骨架的 (φ, ψ) 扭转角的分布,并且是实验结构模型的最佳质量指标之一。通常,验证软件会报告属于“异常值”、“允许”和“偏好”区域的残基数量。虽然“零无法解释的异常值”可以被视为当前的“黄金标准”,但如果不考虑与预期分布的偏差,这可能会产生误导。我们重温了 Ramachandran Z评分 (Rama-Z),这是一个在二十多年前推出但未得到充分利用的质量指标。我们描述了计算晶体学工具箱中 Rama-Z 分数的重新实现,以及用于估计单个模型的不确定性的算法;最终实现在 Phenix 和 PDB-REDO 中可用。我们讨论了 Rama-Z 分数的解释,并主张将其包含在蛋白质数据库提供的验证报告中。我们还主张将其与结构出版物中的异常值/允许/偏好计数一起报告。



























 京公网安备 11010802027423号
京公网安备 11010802027423号