Solid State Nuclear Magnetic Resonance ( IF 1.8 ) Pub Date : 2020-08-27 , DOI: 10.1016/j.ssnmr.2020.101686 John E Kelly 1 , Christine Chrissian 2 , Ruth E Stark 3
|
Many interesting solid-state targets for biological research do not form crystalline structures; these materials include intrinsically disordered proteins, plant biopolymer composites, cell-wall polysaccharides, and soil organic matter. The absence of aligned repeating structural elements and atomic-level rigidity presents hurdles to achieving structural elucidation and obtaining functional insights. We describe strategies for adapting several solid-state NMR methods to determine the molecular structures and compositions of these amorphous biosolids.
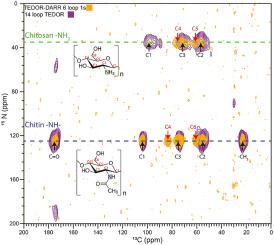
The main spectroscopic problems in studying amorphous structures by NMR are over/under-sampling of the spin signals and spectral complexity. These problems arise in part because amorphous biosolids typically contain a mix of rigid and mobile domains, making it difficult to select a single experiment or set of acquisition conditions that fairly represents all nuclear spins in a carbon-based organic sample. These issues can be addressed by running hybrid experiments, such as using direct excitation alongside cross polarization-based methods, to develop a more holistic picture of the macromolecular system. In situations of spectral crowding or overlap, the structural elucidation strategy can be further assisted by coupling 13C spins to nuclei such as 15N, filtering out portions of the spectrum, highlighting individual moieties of interest, and adding a second or third spectral dimension to an NMR experiment in order to spread out the resonances and link them pairwise through space or through bonds. We discuss practical aspects and illustrations from the recent literature for 1D experiments that use cross or direct polarization and both homo- and heteronuclear 2D and 3D solid-state NMR experiments.
中文翻译:

用于无定形生物固体结构表征的核磁共振实验:实用指南。
许多有趣的生物研究固态靶标并不形成晶体结构;这些材料包括本质上无序的蛋白质、植物生物聚合物复合材料、细胞壁多糖和土壤有机质。缺乏对齐的重复结构元素和原子级的刚性给实现结构阐明和获得功能洞察带来了障碍。我们描述了采用几种固态核磁共振方法来确定这些无定形生物固体的分子结构和组成的策略。
通过核磁共振研究非晶结构的主要光谱问题是自旋信号的过采样/欠采样和光谱复杂性。出现这些问题的部分原因是无定形生物固体通常包含刚性域和移动域的混合,使得很难选择单个实验或一组采集条件来公平地代表碳基有机样品中的所有核自旋。这些问题可以通过运行混合实验来解决,例如使用直接激发和基于交叉极化的方法,以开发大分子系统的更全面的图像。在光谱拥挤或重叠的情况下,可以通过将13 C 自旋耦合到核(例如15 N)、过滤掉光谱的部分、突出显示感兴趣的各个部分以及添加第二个或第三个光谱维度来进一步帮助结构阐明策略。核磁共振实验,以分散共振并通过空间或键将它们成对连接。我们讨论了最近文献中使用交叉或直接极化的 1D 实验以及同核和异核 2D 和 3D 固态 NMR 实验的实际问题和说明。











































 京公网安备 11010802027423号
京公网安备 11010802027423号