当前位置:
X-MOL 学术
›
Microb. Biotechnol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents
Microbial Biotechnology ( IF 4.8 ) Pub Date : 2020-08-25 , DOI: 10.1111/1751-7915.13652 Gen Zou 1, 2 , Meili Xiao 1, 3 , Shunxing Chai 1, 3 , Zhihua Zhu 1, 3 , Ying Wang 2 , Zhihua Zhou 1
Microbial Biotechnology ( IF 4.8 ) Pub Date : 2020-08-25 , DOI: 10.1111/1751-7915.13652 Gen Zou 1, 2 , Meili Xiao 1, 3 , Shunxing Chai 1, 3 , Zhihua Zhu 1, 3 , Ying Wang 2 , Zhihua Zhou 1
Affiliation
|
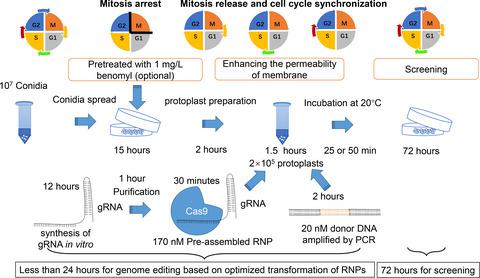
DNA double-strand break (DSB) repair induced by the RNA-programmed nuclease Cas9 has become a popular method for genome editing. Direct genome editing via Cas9-CRISPR gRNA (guide RNA) ribonucleoprotein (RNP) complexes assembled in vitro has also been successful in some fungi. However, the efficiency of direct RNP transformation into fungal protoplasts is currently too low. Here, we report an optimized genome editing approach for filamentous fungi based on RNPs facilitated by adding chemical reagents. We increased the transformation efficiency of RNPs significantly by adding Triton X-100 and prolonging the incubation time, and the editing efficiency reached 100% in Trichoderma reesei and Cordyceps militaris. The optimized RNP-based method also achieved efficient (56.52%) homologous recombination integration with short homology arms (20 bp) and gene disruption (7.37%) that excludes any foreign DNA (selection marker) in T. reesei. In particular, after adding reagents related to mitosis and cell division, the further optimized protocol showed an increased ratio of edited homokaryotic transformants (from 0% to 40.0% for inositol and 71.43% for benomyl) from Aspergillus oryzae, which contains multinucleate spores and protoplasts. Furthermore, the multi-target engineering efficiency of the optimized RNP transformation method was similar to those of methods based on in vivo expression of Cas9. This newly established genome editing system based on RNPs may be widely applicable to construction of genome-edited fungi for the food and medical industries, and has good prospects for commercialization.
中文翻译:

通过化学试剂促进的改进的 CRISPR-Cas9 核糖核蛋白方法对丝状真菌进行有效的基因组编辑
由RNA编程核酸酶Cas9诱导的DNA双链断裂(DSB)修复已成为基因组编辑的流行方法。通过体外组装的 Cas9-CRISPR gRNA(指导 RNA)核糖核蛋白 (RNP) 复合物进行直接基因组编辑也在一些真菌中取得了成功。然而,目前将RNP直接转化为真菌原生质体的效率还太低。在这里,我们报告了一种基于通过添加化学试剂促进的 RNP 的丝状真菌优化基因组编辑方法。我们通过添加Triton X-100并延长孵育时间,显着提高了RNPs的转化效率,在里氏木霉和蛹虫草中的编辑效率均达到100%。基于 RNP 的优化方法还实现了短同源臂 (20 bp) 的高效同源重组整合 (56.52%) 和基因破坏 (7.37%),排除了里氏木霉中的任何外源 DNA(选择标记)。特别是,在添加与有丝分裂和细胞分裂相关的试剂后,进一步优化的方案显示来自米曲霉的编辑同核转化体的比例增加(肌醇从0%增加到40.0%,苯菌灵从71.43%) ,其中含有多核孢子和原生质体。此外,优化后的RNP转化方法的多靶点工程效率与基于Cas9体内表达的方法相似。这种新建立的基于RNP的基因组编辑系统可广泛应用于食品和医疗行业基因组编辑真菌的构建,并具有良好的商业化前景。
更新日期:2020-08-25
中文翻译:

通过化学试剂促进的改进的 CRISPR-Cas9 核糖核蛋白方法对丝状真菌进行有效的基因组编辑
由RNA编程核酸酶Cas9诱导的DNA双链断裂(DSB)修复已成为基因组编辑的流行方法。通过体外组装的 Cas9-CRISPR gRNA(指导 RNA)核糖核蛋白 (RNP) 复合物进行直接基因组编辑也在一些真菌中取得了成功。然而,目前将RNP直接转化为真菌原生质体的效率还太低。在这里,我们报告了一种基于通过添加化学试剂促进的 RNP 的丝状真菌优化基因组编辑方法。我们通过添加Triton X-100并延长孵育时间,显着提高了RNPs的转化效率,在里氏木霉和蛹虫草中的编辑效率均达到100%。基于 RNP 的优化方法还实现了短同源臂 (20 bp) 的高效同源重组整合 (56.52%) 和基因破坏 (7.37%),排除了里氏木霉中的任何外源 DNA(选择标记)。特别是,在添加与有丝分裂和细胞分裂相关的试剂后,进一步优化的方案显示来自米曲霉的编辑同核转化体的比例增加(肌醇从0%增加到40.0%,苯菌灵从71.43%) ,其中含有多核孢子和原生质体。此外,优化后的RNP转化方法的多靶点工程效率与基于Cas9体内表达的方法相似。这种新建立的基于RNP的基因组编辑系统可广泛应用于食品和医疗行业基因组编辑真菌的构建,并具有良好的商业化前景。











































 京公网安备 11010802027423号
京公网安备 11010802027423号