当前位置:
X-MOL 学术
›
J. Biomed. Mater. Res. Part A
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
GSVA score reveals molecular signatures from transcriptomes for biomaterials comparison
Journal of Biomedical Materials Research Part A ( IF 3.9 ) Pub Date : 2020-08-20 , DOI: 10.1002/jbm.a.37090 Marcel R Ferreira 1 , Gerson A Santos 1 , Carlos A Biagi 2 , Wilson A Silva Junior 2 , Willian F Zambuzzi 1
Journal of Biomedical Materials Research Part A ( IF 3.9 ) Pub Date : 2020-08-20 , DOI: 10.1002/jbm.a.37090 Marcel R Ferreira 1 , Gerson A Santos 1 , Carlos A Biagi 2 , Wilson A Silva Junior 2 , Willian F Zambuzzi 1
Affiliation
|
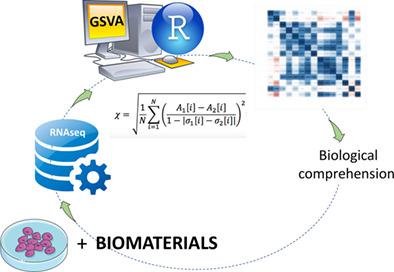
Two in silico methodologies were implemented to reveal the molecular signatures of inorganic hydroxyapatite and β‐TCP materials from a transcriptome database to compare biomaterials. To test this new methodology, we choose the array E‐MTAB‐7219, which contains the transcription profile of osteoblastic cell line seeded onto 15 different biomaterials up to 48 hr. The expansive potential of the methodology was tested from the construction of customized signatures. We present, for the first time, a methodology to compare the performance of different biomaterials using the transcriptome profile of the cell through the Gene set variation analysis (GSVA) score. To test this methodology, we implemented two methods based on MSigDB collections, using all the collections and sub‐collections except the Hallmark collection, which was used in the second method. The result of this analysis provided an initial understanding of biomaterial grouping based on the cell transcriptional landscape. The comparison using GSVA score combined efforts and expand the potential to compare biomaterials using transcriptome profile. Altogether, our results provide a better understanding of the comparison of different biomaterials and suggest a possibility of the new methodology be applied to the prospection of new biomaterials.
中文翻译:

GSVA 评分揭示了用于生物材料比较的转录组的分子特征
实施了两种计算机方法来从转录组数据库中揭示无机羟基磷灰石和 β-TCP 材料的分子特征,以比较生物材料。为了测试这种新方法,我们选择了阵列 E-MTAB-7219,其中包含接种到 15 种不同生物材料上长达 48 小时的成骨细胞系的转录谱。从定制签名的构建中测试了该方法的广泛潜力。我们首次提出了一种方法,通过基因集变异分析 (GSVA) 评分使用细胞的转录组谱来比较不同生物材料的性能。为了测试这种方法,我们实现了两种基于 MSigDB 集合的方法,使用所有集合和子集合,除了在第二种方法中使用的 Hallmark 集合。该分析的结果提供了对基于细胞转录景观的生物材料分组的初步了解。使用 GSVA 分数的比较结合了努力,并扩大了使用转录组谱比较生物材料的潜力。总之,我们的结果提供了对不同生物材料比较的更好理解,并提出了将新方法应用于新生物材料勘探的可能性。
更新日期:2020-08-20
中文翻译:

GSVA 评分揭示了用于生物材料比较的转录组的分子特征
实施了两种计算机方法来从转录组数据库中揭示无机羟基磷灰石和 β-TCP 材料的分子特征,以比较生物材料。为了测试这种新方法,我们选择了阵列 E-MTAB-7219,其中包含接种到 15 种不同生物材料上长达 48 小时的成骨细胞系的转录谱。从定制签名的构建中测试了该方法的广泛潜力。我们首次提出了一种方法,通过基因集变异分析 (GSVA) 评分使用细胞的转录组谱来比较不同生物材料的性能。为了测试这种方法,我们实现了两种基于 MSigDB 集合的方法,使用所有集合和子集合,除了在第二种方法中使用的 Hallmark 集合。该分析的结果提供了对基于细胞转录景观的生物材料分组的初步了解。使用 GSVA 分数的比较结合了努力,并扩大了使用转录组谱比较生物材料的潜力。总之,我们的结果提供了对不同生物材料比较的更好理解,并提出了将新方法应用于新生物材料勘探的可能性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号