Journal of Molecular Graphics and Modelling ( IF 2.7 ) Pub Date : 2020-08-21 , DOI: 10.1016/j.jmgm.2020.107716 Manisha Prajapat 1 , Nishant Shekhar 1 , Phulen Sarma 1 , Pramod Avti 2 , Sanjay Singh 3 , Hardeep Kaur 1 , Anusuya Bhattacharyya 4 , Subodh Kumar 1 , Saurabh Sharma 1 , Ajay Prakash 1 , Bikash Medhi 1
|
Background
The receptor binding domain (RBD) of spike protein S1 domain SARS-CoV-2 plays a key role in the interaction with ACE2, which leads to subsequent S2 domain mediated membrane fusion and incorporation of viral RNA into host cells. In this study we tend to repurpose already approved drugs as inhibitors of the interaction between S1-RBD and the ACE2 receptor.
Methods
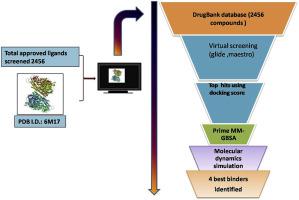
2456 approved drugs were screened against the RBD of S1 protein of SARS-CoV-2 (target PDB ID: 6M17). As the interacting surface between S1-RBD and ACE2 comprises of bigger region, the interacting surface was divided into 3 sites on the basis of interactions (site 1, 2 and 3) and a total of 5 grids were generated (site 1, site 2, site 3, site 1+site 2 and site 2+site 3). A virtual screening was performed using GLIDE implementing HTVS, SP and XP screening. The top hits (on the basis of docking score) were further screened for MM-GBSA. All the top hits were further evaluated in molecular dynamics studies. Performance of the virtual screening protocol was evaluated using enrichment studies.
Result
and discussion: We performed 5 virtual screening against 5 grids generated. A total of 42 compounds were identified after virtual screening. These drugs were further assessed for their interaction dynamics in molecular dynamics simulation. On the basis of molecular dynamics studies, we come up with 10 molecules with favourable interaction profile, which also interacted with physiologically important residues (residues taking part in the interaction between S1-RBD and ACE2. These are antidiabetic (acarbose), vitamins (riboflavin and levomefolic acid), anti-platelet agents (cangrelor), aminoglycoside antibiotics (Kanamycin, amikacin) bronchodilator (fenoterol), immunomodulator (lamivudine), and anti-neoplastic agents (mitoxantrone and vidarabine). However, while considering the relative side chain fluctuations when compared to the S1-RBD: ACE2 complex riboflavin, fenoterol, cangrelor and vidarabine emerged out as molecules with prolonged relative stability.
Conclusion
We identified 4 already approved drugs (riboflavin, fenoterol, cangrelor and vidarabine) as possible agents for repurposing as inhibitors of S1:ACE2 interaction. In-vitro validation of these findings are necessary for identification of a safe and effective inhibitor of S1: ACE2 mediated entry of SARS-CoV-2 into the host cell.
中文翻译:

已批准药物作为 SARS-CoV-2 中刺突蛋白 S1 结构域和 ACE2 相互作用抑制剂的虚拟筛选和分子动力学研究。
背景
SARS-CoV-2 刺突蛋白 S1 结构域的受体结合结构域 (RBD) 在与 ACE2 相互作用中发挥关键作用,从而导致随后的 S2 结构域介导的膜融合以及病毒 RNA 掺入宿主细胞。在这项研究中,我们倾向于将已批准的药物重新用作 S1-RBD 和 ACE2 受体之间相互作用的抑制剂。
方法
针对 SARS-CoV-2 S1 蛋白的 RBD 筛选了 2456 种已批准药物(目标 PDB ID:6M17)。由于S1-RBD和ACE2之间的相互作用表面包含较大的区域,因此根据相互作用将相互作用表面分为3个位点(位点1、2和3),总共生成5个网格(位点1、位点2) 、站点 3、站点 1+站点 2 和站点 2+站点 3)。使用 GLIDE 实施 HTVS、SP 和 XP 筛选进行虚拟筛选。进一步筛选 MM-GBSA 的热门命中(基于对接分数)。所有热门歌曲都在分子动力学研究中得到了进一步评估。使用富集研究评估虚拟筛选方案的性能。
结果
和讨论:我们对生成的 5 个网格进行了 5 次虚拟筛选。经过虚拟筛选,共鉴定出 42 种化合物。在分子动力学模拟中进一步评估了这些药物的相互作用动力学。在分子动力学研究的基础上,我们提出了 10 个具有良好相互作用特征的分子,它们也与生理上重要的残基(参与 S1-RBD 和 ACE2 之间相互作用的残基)相互作用。这些是抗糖尿病药(阿卡波糖)、维生素(核黄素)和左美叶酸)、抗血小板药物(坎格瑞洛)、氨基糖苷类抗生素(卡那霉素、阿米卡星)、支气管扩张剂(非诺特罗)、免疫调节剂(拉米夫定)和抗肿瘤药物(米托蒽醌和阿糖腺苷)。然而,在考虑相对侧链波动的同时与 S1-RBD 相比:ACE2 复合物核黄素、非诺特罗、坎格雷洛和阿糖腺苷作为具有长期相对稳定性的分子出现。
结论
我们确定了 4 种已批准的药物(核黄素、非诺特罗、坎格雷洛和阿糖腺苷)作为重新用作 S1:ACE2 相互作用抑制剂的可能药物。这些发现的体外验证对于鉴定 S1:ACE2 介导的 SARS-CoV-2 进入宿主细胞的安全有效抑制剂是必要的。











































 京公网安备 11010802027423号
京公网安备 11010802027423号