当前位置:
X-MOL 学术
›
RSC Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A thorough analysis and categorization of bacterial interrupted adenylation domains, including previously unidentified families
RSC Chemical Biology ( IF 4.2 ) Pub Date : 2020-08-18 , DOI: 10.1039/d0cb00092b Taylor A Lundy 1 , Shogo Mori 1 , Sylvie Garneau-Tsodikova 1
RSC Chemical Biology ( IF 4.2 ) Pub Date : 2020-08-18 , DOI: 10.1039/d0cb00092b Taylor A Lundy 1 , Shogo Mori 1 , Sylvie Garneau-Tsodikova 1
Affiliation
|
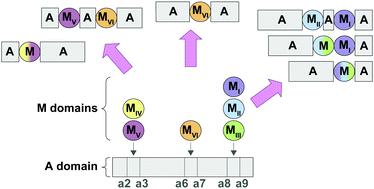
Interrupted adenylation (A) domains are key to the immense structural diversity seen in the nonribosomal peptide (NRP) class of natural products (NPs). Interrupted A domains are A domains that contain within them the catalytic portion of another domain, most commonly a methylation (M) domain. It has been well documented that methylation events occur with extreme specificity on either the backbone (N-) or side chain (O- or S-) of the amino acid (or amino acid-like) building blocks of NRPs. Here, through taxonomic and phylogenetic analyses as well as multiple sequence alignments, we evaluated the similarities and differences between interrupted A domains. We probed their taxonomic distribution amongst bacterial organisms, their evolutionary relatedness, and described conserved motifs of each type of M domain found to be embedded in interrupted A domains. Additionally, we categorized interrupted A domains and the M domains within them into a total of seven distinct families and six different types, respectively. The families of interrupted A domains include two new families, 6 and 7, that possess new architectures. Rather than being interrupted between the previously described a2–a3 or a8–a9 of the ten conserved A domain sequence motifs (a1–a10), family 6 contains an M domain between a6–a7, a previously unknown interruption site. Family 7 demonstrates that di-interrupted A domains exist in Nature, containing an M domain between a2–a3 as well as one between a6–a7, displaying a novel arrangement. These in-depth investigations of amino acid sequences deposited in the NCBI database highlighted the prevalence of interrupted A domains in bacterial organisms, with each family of interrupted A domains having a different taxonomic distribution. They also emphasized the importance of utilizing a broad range of bacteria for NP discovery. Categorization of the families of interrupted A domains and types of M domains allowed for a better understanding of the trends of naturally occurring interrupted A domains, which illuminated patterns and insights on how to harness them for future engineering studies.
中文翻译:

对细菌间断腺苷酸化结构域的彻底分析和分类,包括以前未鉴定的家族
中断的腺苷酸化 (A) 结构域是非核糖体肽 (NRP) 类天然产物 (NP) 中巨大结构多样性的关键。中断的 A 域是其中包含另一个域的催化部分的 A 域,最常见的是甲基化 (M) 域。已有充分证明,甲基化事件在主链 ( N -) 或侧链 ( O - 或S-) NRP 的氨基酸(或氨基酸样)构建块。在这里,通过分类学和系统发育分析以及多序列比对,我们评估了中断的 A 域之间的异同。我们探讨了它们在细菌生物体中的分类分布、它们的进化相关性,并描述了发现嵌入中断 A 域的每种类型 M 域的保守基序。此外,我们将中断的 A 域和其中的 M 域分别分为七个不同的家族和六种不同的类型。中断的 A 域的家族包括两个新的家族,6 和 7,它们拥有新的架构。而不是在先前描述的 10 个保守 A 域序列基序 (a1-a10) 的 a2-a3 或 a8-a9 之间中断,家族 6 在 a6-a7 之间包含一个 M 域,这是一个以前未知的中断位点。家族 7 表明自然中存在双中断的 A 域,包含 a2-a3 之间的 M 域以及 a6-a7 之间的一个域,显示出一种新颖的排列。这些对保存在 NCBI 数据库中的氨基酸序列的深入研究突出了细菌生物体中 A 结构域中断的普遍性,每个 A 结构域的中断家族具有不同的分类分布。他们还强调了利用广泛的细菌进行 NP 发现的重要性。对中断的 A 域家族和 M 域的类型进行分类,可以更好地了解自然发生的中断 A 域的趋势,
更新日期:2020-10-08
中文翻译:

对细菌间断腺苷酸化结构域的彻底分析和分类,包括以前未鉴定的家族
中断的腺苷酸化 (A) 结构域是非核糖体肽 (NRP) 类天然产物 (NP) 中巨大结构多样性的关键。中断的 A 域是其中包含另一个域的催化部分的 A 域,最常见的是甲基化 (M) 域。已有充分证明,甲基化事件在主链 ( N -) 或侧链 ( O - 或S-) NRP 的氨基酸(或氨基酸样)构建块。在这里,通过分类学和系统发育分析以及多序列比对,我们评估了中断的 A 域之间的异同。我们探讨了它们在细菌生物体中的分类分布、它们的进化相关性,并描述了发现嵌入中断 A 域的每种类型 M 域的保守基序。此外,我们将中断的 A 域和其中的 M 域分别分为七个不同的家族和六种不同的类型。中断的 A 域的家族包括两个新的家族,6 和 7,它们拥有新的架构。而不是在先前描述的 10 个保守 A 域序列基序 (a1-a10) 的 a2-a3 或 a8-a9 之间中断,家族 6 在 a6-a7 之间包含一个 M 域,这是一个以前未知的中断位点。家族 7 表明自然中存在双中断的 A 域,包含 a2-a3 之间的 M 域以及 a6-a7 之间的一个域,显示出一种新颖的排列。这些对保存在 NCBI 数据库中的氨基酸序列的深入研究突出了细菌生物体中 A 结构域中断的普遍性,每个 A 结构域的中断家族具有不同的分类分布。他们还强调了利用广泛的细菌进行 NP 发现的重要性。对中断的 A 域家族和 M 域的类型进行分类,可以更好地了解自然发生的中断 A 域的趋势,











































 京公网安备 11010802027423号
京公网安备 11010802027423号