当前位置:
X-MOL 学术
›
Mol. Plant Pathol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
EffectorK, a comprehensive resource to mine for Ralstonia, Xanthomonas, and other published effector interactors in the Arabidopsis proteome
Molecular Plant Pathology ( IF 4.8 ) Pub Date : 2020-08-15 , DOI: 10.1111/mpp.12965 Manuel González-Fuente 1 , Sébastien Carrère 1 , Dario Monachello 2, 3 , Benjamin G Marsella 4 , Anne-Claire Cazalé 1 , Claudine Zischek 1 , Raka M Mitra 5 , Nathalie Rezé 2, 3 , Ludovic Cottret 1 , M Shahid Mukhtar 4 , Claire Lurin 2, 3 , Laurent D Noël 1 , Nemo Peeters 1
Molecular Plant Pathology ( IF 4.8 ) Pub Date : 2020-08-15 , DOI: 10.1111/mpp.12965 Manuel González-Fuente 1 , Sébastien Carrère 1 , Dario Monachello 2, 3 , Benjamin G Marsella 4 , Anne-Claire Cazalé 1 , Claudine Zischek 1 , Raka M Mitra 5 , Nathalie Rezé 2, 3 , Ludovic Cottret 1 , M Shahid Mukhtar 4 , Claire Lurin 2, 3 , Laurent D Noël 1 , Nemo Peeters 1
Affiliation
|
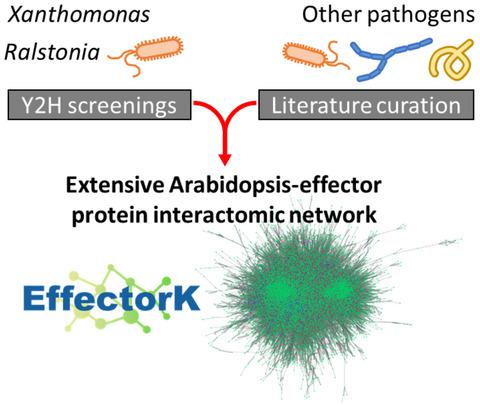
Pathogens deploy effector proteins that interact with host proteins to manipulate the host physiology to the pathogen's own benefit. However, effectors can also be recognized by host immune proteins, leading to the activation of defence responses. Effectors are thus essential components in determining the outcome of plant–pathogen interactions. Despite major efforts to decipher effector functions, our current knowledge on effector biology is scattered and often limited. In this study, we conducted two systematic large‐scale yeast two‐hybrid screenings to detect interactions between Arabidopsis thaliana proteins and effectors from two vascular bacterial pathogens: Ralstonia pseudosolanacearum and Xanthomonas campestris. We then constructed an interactomic network focused on Arabidopsis and effector proteins from a wide variety of bacterial, oomycete, fungal, and invertebrate pathogens. This network contains our experimental data and protein–protein interactions from 2,035 peer‐reviewed publications (48,200 Arabidopsis–Arabidopsis and 1,300 Arabidopsis–effector protein interactions). Our results show that effectors from different species interact with both common and specific Arabidopsis interactors, suggesting dual roles as modulators of generic and adaptive host processes. Network analyses revealed that effector interactors, particularly “effector hubs” and bacterial core effector interactors, occupy important positions for network organization, as shown by their larger number of protein interactions and centrality. These interactomic data were incorporated in EffectorK, a new graph‐oriented knowledge database that allows users to navigate the network, search for homology, or find possible paths between host and/or effector proteins. EffectorK is available at www.effectork.org and allows users to submit their own interactomic data.
中文翻译:

EffectorK,一种综合资源,可用于挖掘拟南芥蛋白质组中的 Ralstonia、Xanthomonas 和其他已发表的效应子相互作用因子
病原体利用与宿主蛋白质相互作用的效应蛋白来操纵宿主生理机能,从而为病原体自身谋利。然而,效应子也可以被宿主免疫蛋白识别,从而激活防御反应。因此,效应子是决定植物与病原体相互作用结果的重要组成部分。尽管我们在破译效应器功能方面付出了巨大努力,但我们目前对效应器生物学的了解仍然分散且往往有限。在这项研究中,我们进行了两次系统的大规模酵母双杂交筛选,以检测拟南芥蛋白与两种维管细菌病原体(拟茄青菌和野油菜黄单胞菌)效应子之间的相互作用。然后,我们构建了一个相互作用组学网络,重点关注拟南芥和来自多种细菌、卵菌、真菌和无脊椎动物病原体的效应蛋白。该网络包含我们的实验数据和来自 2,035 个同行评审出版物的蛋白质-蛋白质相互作用(48,200 个拟南芥-拟南芥和 1,300 个拟南芥-效应蛋白相互作用)。我们的结果表明,来自不同物种的效应子与常见和特定的拟南芥相互作用子相互作用,表明其作为通用和适应性宿主过程调节剂的双重作用。网络分析表明,效应器相互作用体,特别是“效应器中心”和细菌核心效应器相互作用器,在网络组织中占据重要地位,正如它们大量的蛋白质相互作用和中心性所表明的那样。 这些相互作用组学数据被纳入 EffectorK 中,这是一个新的面向图形的知识数据库,允许用户导航网络、搜索同源性或找到宿主和/或效应蛋白之间的可能路径。 EffectorK 可在 www.effectork.org 上获取,并允许用户提交自己的交互组学数据。
更新日期:2020-08-15
中文翻译:

EffectorK,一种综合资源,可用于挖掘拟南芥蛋白质组中的 Ralstonia、Xanthomonas 和其他已发表的效应子相互作用因子
病原体利用与宿主蛋白质相互作用的效应蛋白来操纵宿主生理机能,从而为病原体自身谋利。然而,效应子也可以被宿主免疫蛋白识别,从而激活防御反应。因此,效应子是决定植物与病原体相互作用结果的重要组成部分。尽管我们在破译效应器功能方面付出了巨大努力,但我们目前对效应器生物学的了解仍然分散且往往有限。在这项研究中,我们进行了两次系统的大规模酵母双杂交筛选,以检测拟南芥蛋白与两种维管细菌病原体(拟茄青菌和野油菜黄单胞菌)效应子之间的相互作用。然后,我们构建了一个相互作用组学网络,重点关注拟南芥和来自多种细菌、卵菌、真菌和无脊椎动物病原体的效应蛋白。该网络包含我们的实验数据和来自 2,035 个同行评审出版物的蛋白质-蛋白质相互作用(48,200 个拟南芥-拟南芥和 1,300 个拟南芥-效应蛋白相互作用)。我们的结果表明,来自不同物种的效应子与常见和特定的拟南芥相互作用子相互作用,表明其作为通用和适应性宿主过程调节剂的双重作用。网络分析表明,效应器相互作用体,特别是“效应器中心”和细菌核心效应器相互作用器,在网络组织中占据重要地位,正如它们大量的蛋白质相互作用和中心性所表明的那样。 这些相互作用组学数据被纳入 EffectorK 中,这是一个新的面向图形的知识数据库,允许用户导航网络、搜索同源性或找到宿主和/或效应蛋白之间的可能路径。 EffectorK 可在 www.effectork.org 上获取,并允许用户提交自己的交互组学数据。











































 京公网安备 11010802027423号
京公网安备 11010802027423号