Archives of Biochemistry and Biophysics ( IF 3.8 ) Pub Date : 2020-08-15 , DOI: 10.1016/j.abb.2020.108545 Tyler Eck 1 , Seema Patel 1 , Thomas Candela 1 , Katherine Leon H 1 , Michael Little 1 , Natalia E Reis 1 , Uththara Liyanagunawardana 1 , Ueli Gubler 1 , Cheryl A Janson 1 , Jaclyn Catalano 1 , Nina M Goodey 1
|
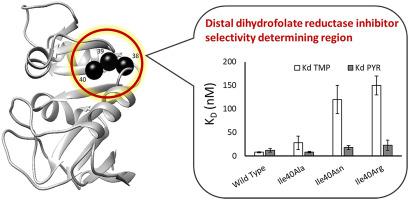
Many antibacterial and antiparasitic drugs work by competitively inhibiting dihydrofolate reductase (DHFR), a vital enzyme in folate metabolism. The interactions between inhibitors and DHFR active site residues are known in many homologs but the contributions from distal residues are less understood. Identifying distal residues that aid in inhibitor binding can improve targeted drug development programs by accounting for distant influences that may be less conserved and subject to frequent resistance causing mutations. Previously, a novel, homology-based, computational approach that mines ligand inhibition data was used to predict residues involved in inhibitor selectivity in the DHFR family. Expectedly, some inhibitor selectivity determining residue positions were predicted to lie in the active site and coincide with experimentally known inhibitor selectivity determining positions. However, other residues that group spatially in clusters distal to the active site have not been previously investigated. In this study, the effect of introducing amino acid substitutions at one of these predicted clusters (His38-Ala39-Ile40) on the inhibitor selectivity profile in Bacillus stearothermophilus dihydrofolate reductase (Bs DHFR) was investigated. Mutations were introduced into these cluster positions to change sidechain chemistry and size. We determined kcat and KM values and measured KD values at equilibrium for two competitive DHFR inhibitors, trimethoprim (TMP) and pyrimethamine (PYR). Mutations in the His38-Ala39-Ile40 cluster significantly impacted inhibitor binding and TMP/PYR selectivity - seven out of nine mutations resulted in tighter binding to PYR when compared to TMP. These data suggest that the His38-Ala39-Ile40 cluster is a distal inhibitor selectivity determining region that favors PYR binding in Bs DHFR and, possibly, throughout the DHFR family.
中文翻译:

突变分析证实嗜热脂肪芽孢杆菌二氢叶酸还原酶中存在远端抑制剂选择性决定残基。
许多抗菌和抗寄生虫药物通过竞争性抑制二氢叶酸还原酶 (DHFR) 发挥作用,二氢叶酸还原酶是叶酸代谢中的重要酶。在许多同系物中,抑制剂和 DHFR 活性位点残基之间的相互作用是已知的,但远端残基的贡献却知之甚少。识别有助于抑制剂结合的远端残基可以通过考虑可能不太保守且经常遭受引起耐药性突变的远端影响来改善靶向药物开发计划。此前,一种新颖的、基于同源性的计算方法挖掘配体抑制数据,用于预测 DHFR 家族中参与抑制剂选择性的残基。预期,一些抑制剂选择性决定残基位置被预测位于活性位点并且与实验已知的抑制剂选择性决定位置一致。然而,之前尚未研究过在空间上聚集在远离活性位点的簇中的其他残基。在本研究中,研究了在这些预测簇之一 (His38-Ala39-Ile40) 中引入氨基酸取代对嗜热脂肪芽孢杆菌二氢叶酸还原酶 ( Bs DHFR) 抑制剂选择性谱的影响。将突变引入这些簇位置以改变侧链化学和大小。我们确定了k cat和 K M值,并测量了两种竞争性 DHFR 抑制剂甲氧苄啶 (TMP) 和乙胺嘧啶 (PYR) 平衡时的 K D值。 His38-Ala39-Ile40 簇中的突变显着影响抑制剂结合和 TMP/PYR 选择性 - 与 TMP 相比,九个突变中的七个导致与 PYR 的结合更紧密。 这些数据表明 His38-Ala39-Ile40 簇是一个远端抑制剂选择性决定区域,有利于Bs DHFR 中的 PYR 结合,并且可能在整个 DHFR 家族中。











































 京公网安备 11010802027423号
京公网安备 11010802027423号